Proteomics reveals the role of the EamB transporter from Aeromonas hydrophila LP-2 in biofilm formation
IF 2.8
2区 生物学
Q2 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
Abstract
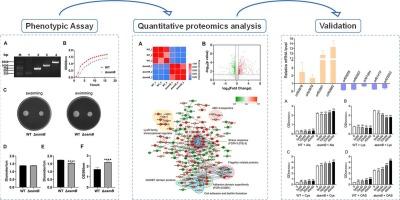
Biofilms play a pivotal role in the survival and persistence of microorganisms, endowing them with heightened resistance to environmental stressors and antimicrobial agents. The EamB protein, which encodes an inner membrane transporter, acted as a negative regulator of biofilm formation, and the gene eamB deletion in the pathogen Aeromonas hydrophila LP-2 resulted in a significant increase in biofilm formation. Proteomic analysis revealed a total of 616 differentially abundant proteins between the ΔeamB and wild-type (WT) strains, with 308 downregulated and 308 upregulated. RT-qPCR was employed to verify the stability and accuracy of the proteomics data. Bioinformatic analysis indicated that EamB is involved in critical bacterial biological processes, including flagellar assembly, amino acid metabolism, and fatty acid degradation. Biofilm formation assays further revealed that supplementation with exogenous lysine significantly inhibited biofilm formation in the ΔeamB strain, conversely, exogenous cysteine and O-acetylserine obviously increased biofilm formation in the ΔeamB strain. These findings demonstrated that EamB may modulate bacterial biofilm formation in A. hydrophila through the regulation of amino acid metabolism. This finding provides novel insights into the regulatory mechanism underlying biofilm formation and highlights potential targets for the development of future antibacterial strategies.
Significance statement
This study elucidates the critical role of the eamB gene in Aeromonas hydrophila, a significant aquatic pathogen, by demonstrating its impact on biofilm formation and physiological traits. Through comparative proteomic analysis, we identified 616 differentially abundant proteins in the ΔeamB mutant, revealing its involvement in key metabolic pathways such as amino acid metabolism, flagellar assembly, and fatty acid degradation. Notably, eamB deletion enhanced biofilm formation, while exogenous amino acids like cysteine and O-acetylserine obviously increased biofilm formation in the ΔeamB strain. These findings highlight EamB as a regulator of biofilm formation, offering novel molecular insights into bacterial pathogenicity. This research advances our understanding of biofilm-associated antibiotic resistance and provides potential targets for developing strategies to mitigate infections caused by A. hydrophila in aquaculture and public health.
蛋白质组学揭示了来自嗜水气单胞菌LP-2的EamB转运体在生物膜形成中的作用。
生物膜在微生物的生存和持久性中起着关键作用,赋予它们对环境压力和抗菌剂的更高抵抗力。编码一种内膜转运蛋白的EamB蛋白是生物膜形成的负调控因子,病原嗜水气单胞菌LP-2中EamB基因的缺失导致生物膜形成的显著增加。蛋白质组学分析显示,ΔeamB与野生型(WT)菌株之间共有616个差异丰富的蛋白,其中308个下调,308个上调。采用RT-qPCR验证蛋白质组学数据的稳定性和准确性。生物信息学分析表明,EamB参与了关键的细菌生物学过程,包括鞭毛组装,氨基酸代谢和脂肪酸降解。生物膜形成实验进一步表明,添加外源赖氨酸显著抑制了ΔeamB菌株的生物膜形成,相反,外源半胱氨酸和o -乙酰丝氨酸明显增加了ΔeamB菌株的生物膜形成。这些结果表明,EamB可能通过调节氨基酸代谢来调节嗜水单胞菌生物膜的形成。这一发现为生物膜形成的调控机制提供了新的见解,并为未来抗菌策略的发展提供了潜在的靶点。意义声明:本研究通过证明eamB基因对生物膜形成和生理特性的影响,阐明了eamB基因在嗜水气单胞菌(一种重要的水生病原体)中的关键作用。通过比较蛋白质组学分析,我们在ΔeamB突变体中鉴定了616个差异表达蛋白,揭示了其参与氨基酸代谢、鞭毛组装和脂肪酸降解等关键代谢途径。值得注意的是,eamB的缺失促进了生物膜的形成,而外源氨基酸如半胱氨酸和o -乙酰丝氨酸则明显促进了ΔeamB菌株生物膜的形成。这些发现强调了EamB作为生物膜形成的调节剂,为细菌致病性提供了新的分子见解。该研究促进了我们对生物膜相关抗生素耐药性的理解,并为制定减轻水产养殖和公共卫生中嗜水单胞杆菌感染的策略提供了潜在的目标。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of proteomics
生物-生化研究方法
CiteScore
7.10
自引率
3.00%
发文量
227
审稿时长
73 days
期刊介绍:
Journal of Proteomics is aimed at protein scientists and analytical chemists in the field of proteomics, biomarker discovery, protein analytics, plant proteomics, microbial and animal proteomics, human studies, tissue imaging by mass spectrometry, non-conventional and non-model organism proteomics, and protein bioinformatics. The journal welcomes papers in new and upcoming areas such as metabolomics, genomics, systems biology, toxicogenomics, pharmacoproteomics.
Journal of Proteomics unifies both fundamental scientists and clinicians, and includes translational research. Suggestions for reviews, webinars and thematic issues are welcome.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


