Human miRNA–disease Association Prediction Via Residual GraphSAGE With Nonlinear Adaptive Feature Fusion and Triplet Contrastive Learning
IF 4.5
2区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
MicroRNAs (miRNAs) play pivotal roles in cellular regulation, and their dysregulation is closely linked to a wide spectrum of human diseases; thus, accurate miRNA–disease association prediction is critical for guiding experimental validation and therapeutic development. In this work, we propose RGFMDA, an innovative framework designed to predict miRNA-disease associations more effectively. RGFMDA employs a residual graph sampling and aggregation network to enhance information localization within miRNA and disease networks. It also features a nonlinear integration of features and a global context integration module that synergistically combine feature interactions and oversee global dependencies. Additionally, the framework uses triplet contrastive learning to refine the distinction between associated and non-associated miRNA-disease pairs, enhancing the accuracy of predictions. On the HMDD v2.0 benchmark, RGFMDA achieved an AUC of 0.9524, surpassing existing approaches whose reported AUC values range from approximately 0.916 to 0.942. On the HMDD v3.2 dataset, RGFMDA further improved performance with an AUC of 0.9604, exceeding state-of-the-art models that demonstrate AUCs between roughly 0.912 and 0.953. Case studies involving lung, esophageal, breast, and colorectal cancers have further confirmed the efficacy of RGFMDA. In summary, RGFMDA represents a robust and reliable computational tool for uncovering novel miRNA–disease associations, thereby facilitating future biological discovery and therapeutic development.
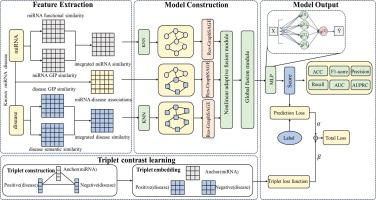
基于非线性自适应特征融合和三联体对比学习的残差GraphSAGE人类mirna -疾病关联预测。
MicroRNAs (miRNAs)在细胞调控中发挥关键作用,其失调与广泛的人类疾病密切相关;因此,准确的mirna -疾病关联预测对于指导实验验证和治疗开发至关重要。在这项工作中,我们提出了RGFMDA,这是一个创新的框架,旨在更有效地预测mirna与疾病的关联。RGFMDA采用残差图采样和聚合网络来增强miRNA和疾病网络中的信息定位。它还具有非线性特性集成和全局上下文集成模块,可以协同地组合特性交互并监督全局依赖关系。此外,该框架使用三联体对比学习来细化相关和非相关mirna -疾病对之间的区别,提高预测的准确性。在HMDD v2.0基准上,RGFMDA实现了0.9524的AUC,超过了现有的方法,其报告的AUC值范围约为0.916到0.942。在HMDD v3.2数据集上,RGFMDA进一步提高了性能,AUC为0.9604,超过了最先进的模型,AUC大约在0.912到0.953之间。涉及肺癌、食管癌、乳腺癌和结直肠癌的病例研究进一步证实了RGFMDA的疗效。总之,RGFMDA是一个强大而可靠的计算工具,用于发现新的mirna -疾病关联,从而促进未来的生物学发现和治疗开发。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Molecular Biology
生物-生化与分子生物学
CiteScore
11.30
自引率
1.80%
发文量
412
审稿时长
28 days
期刊介绍:
Journal of Molecular Biology (JMB) provides high quality, comprehensive and broad coverage in all areas of molecular biology. The journal publishes original scientific research papers that provide mechanistic and functional insights and report a significant advance to the field. The journal encourages the submission of multidisciplinary studies that use complementary experimental and computational approaches to address challenging biological questions.
Research areas include but are not limited to: Biomolecular interactions, signaling networks, systems biology; Cell cycle, cell growth, cell differentiation; Cell death, autophagy; Cell signaling and regulation; Chemical biology; Computational biology, in combination with experimental studies; DNA replication, repair, and recombination; Development, regenerative biology, mechanistic and functional studies of stem cells; Epigenetics, chromatin structure and function; Gene expression; Membrane processes, cell surface proteins and cell-cell interactions; Methodological advances, both experimental and theoretical, including databases; Microbiology, virology, and interactions with the host or environment; Microbiota mechanistic and functional studies; Nuclear organization; Post-translational modifications, proteomics; Processing and function of biologically important macromolecules and complexes; Molecular basis of disease; RNA processing, structure and functions of non-coding RNAs, transcription; Sorting, spatiotemporal organization, trafficking; Structural biology; Synthetic biology; Translation, protein folding, chaperones, protein degradation and quality control.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


