Semantic consistency-guided patch-wise relation graph reasoning scheme for lung cancer organoid segmentation in brightfield microscopy
IF 4.8
2区 医学
Q1 COMPUTER SCIENCE, INTERDISCIPLINARY APPLICATIONS
引用次数: 0
Abstract
Background and Objective
: Patient-derived organoids (PDOs) have recently opened new possibilities for personalized precision medicine. Specifically, the segmentation of live PDOs in brightfield microscopy is essential for organoid morphological evaluation, cell viability assays, and drug screening. However, accurately and automatically annotating live organoids remains challenging due to factors such as low contrast, varying organoid characteristics, imaging artifacts, and the scarcity of large-scale annotated datasets. This study aims to address these challenges by proposing an advanced method for live PDOs segmentation even with limited training data.
Methods:
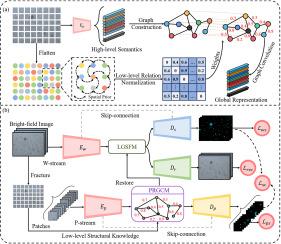
We propose a semantic consistency-guided patch-wise relation graph reasoning scheme, which allows local–global semantics extraction, multi-task-based representation refinement and semantic consistency constraint for live PDOs segmentation. Specifically, our approach consists of two input-specific streams designed to extract high-level whole-image local and patch-wise semantic representations. Then a patch-wise relation graph convolution module (PRGCM) is devised to exploit global prior morphological properties by integrating low-level spatial relatedness into patch-wise high-level semantic representations using graph convolution. Subsequently, a local–global semantics fusion module (LGSFM) is deployed to enable local–global contextual semantic fusion. In the decoding stage, we develop two auxiliary learning tasks, with a patch segmentation decoder to guarantee semantic homogeneity by a novel loss function, called stream consistency (SC) loss, along with a reconstruction decoder to capture powerful and discriminative representation without additional annotations.
Results:
We introduce LiveOrganoid, a large-scale dataset of manually annotated lung cancer brightfield images, consisting of lung PDOs with diverse morphologies. Our scheme demonstrates superior performance on LiveOrganoid, achieving state-of-the-art results compared to recent methods, even with fewer training data. Additionally, we evaluate our method’s generalization for different imaging modality and other cell-culture segmentation tasks, including cell and nuclear segmentation in tissue images.
Conclusions:
Our proposed semantic consistency-guided patchwise relation graph reasoning scheme addresses the challenges in live PDOs segmentation, paving the way for improved organoid morphological evaluation, cell viability assays, and drug screening. The method’s generalizability to other cell-culture segmentation tasks highlights its potential for broader applications in precision medicine.
语义一致性引导的亮场显微镜肺癌类器官分割斑块关系图推理方案
背景与目的:患者源性类器官(PDOs)最近为个性化精准医疗开辟了新的可能性。具体来说,在明场显微镜下,活体PDOs的分割对于类器官形态评估、细胞活力测定和药物筛选至关重要。然而,由于低对比度、不同的类器官特征、成像伪影以及大规模注释数据集的稀缺性等因素,准确、自动地注释活体类器官仍然具有挑战性。本研究旨在通过提出一种先进的实时pdo分割方法来解决这些挑战,即使训练数据有限。方法:提出了一种基于语义一致性的补丁关系图推理方案,该方案支持局部全局语义提取、基于多任务的表示细化和语义一致性约束。具体来说,我们的方法包括两个特定于输入的流,用于提取高级整体图像局部和补丁语义表示。在此基础上,设计了一种基于补丁的关系图卷积模块(PRGCM),通过图卷积将低层次空间相关性集成到补丁高级语义表示中,从而利用全局先验形态学特性。随后,部署本地-全局语义融合模块(LGSFM)实现本地-全局上下文语义融合。在解码阶段,我们开发了两个辅助学习任务,一个是补丁分割解码器,通过一种称为流一致性(SC)损失的新型损失函数来保证语义同质性,另一个是重建解码器,在没有额外注释的情况下捕获强大的判别表示。结果:我们引入了LiveOrganoid,这是一个人工注释肺癌亮场图像的大规模数据集,由不同形态的肺pdo组成。我们的方案在LiveOrganoid上展示了卓越的性能,与最近的方法相比,即使使用更少的训练数据,也能获得最先进的结果。此外,我们还评估了我们的方法在不同成像模式和其他细胞培养分割任务中的泛化性,包括组织图像中的细胞和核分割。结论:我们提出的语义一致性引导的补丁关系图推理方案解决了活体pdo分割中的挑战,为改进类器官形态学评估、细胞活力测定和药物筛选铺平了道路。该方法可推广到其他细胞培养分割任务,突出了其在精密医学中更广泛应用的潜力。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Computer methods and programs in biomedicine
工程技术-工程:生物医学
CiteScore
12.30
自引率
6.60%
发文量
601
审稿时长
135 days
期刊介绍:
To encourage the development of formal computing methods, and their application in biomedical research and medical practice, by illustration of fundamental principles in biomedical informatics research; to stimulate basic research into application software design; to report the state of research of biomedical information processing projects; to report new computer methodologies applied in biomedical areas; the eventual distribution of demonstrable software to avoid duplication of effort; to provide a forum for discussion and improvement of existing software; to optimize contact between national organizations and regional user groups by promoting an international exchange of information on formal methods, standards and software in biomedicine.
Computer Methods and Programs in Biomedicine covers computing methodology and software systems derived from computing science for implementation in all aspects of biomedical research and medical practice. It is designed to serve: biochemists; biologists; geneticists; immunologists; neuroscientists; pharmacologists; toxicologists; clinicians; epidemiologists; psychiatrists; psychologists; cardiologists; chemists; (radio)physicists; computer scientists; programmers and systems analysts; biomedical, clinical, electrical and other engineers; teachers of medical informatics and users of educational software.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


