PPARγ acetylation governs mammary adenocarcinoma tumor growth via acetylated residues that determine DNA sequence-specific binding
IF 7.3
1区 医学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
Peroxisome proliferator-activated receptor γ (PPARγ), which is expressed in a variety of malignancies, governs biological functions through transcriptional programs. Defining the molecular mechanisms governing the selection of canonical versus non-canonical PPARγ binding sequences may provide the opportunity to design regulators with distinct functions and side effects. Acetylation at K268/293 in mouse Pparγ2 participates in the regulation of adipose tissue differentiation, and the conserved lysine residues (K154/155) in mouse Pparγ1 governs lipogenesis in breast cancer cells. Herein, the PPARγ1 acetylated residues K154/155 were shown to be essential for oncogenic ErbB2 driven breast cancer growth and mammary tumor stem cell expansion in vivo. The induction of transcriptional modules governing growth factor signaling, lipogenesis, cellular apoptosis, and stem cell expansion were dependent upon K154/155. The acetylation status of the K154/155 residues determined the selection of genome-wide DNA binding sites, altering the selection from canonical to non-canonical (C/EBP) DNA sequence-specific binding. The gene signature reflecting the acetylation-dependent genomic occupancy in lipogenesis provided predictive value in survival outcomes of ErbB2+ breast cancer. The Pparγ1 acetylation site is critical for ErbB2-induced breast cancer tumor growth and may represent a relevant target for therapeutic coextinction.
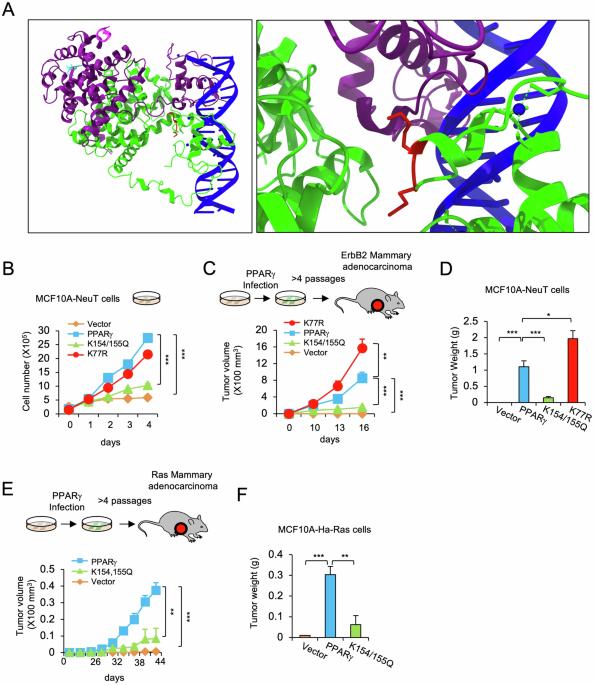
PPARγ乙酰化通过决定DNA序列特异性结合的乙酰化残基控制乳腺腺癌肿瘤的生长。
过氧化物酶体增殖物激活受体γ (PPARγ)在多种恶性肿瘤中表达,通过转录程序控制生物学功能。明确规范与非规范PPARγ结合序列选择的分子机制可能为设计具有不同功能和副作用的调节因子提供机会。小鼠Pparγ2中K268/293位点的乙酰化参与了脂肪组织分化的调控,小鼠Pparγ1中保守的赖氨酸残基(K154/155)调控了乳腺癌细胞的脂肪生成。在本研究中,ppar γ - 1乙酰化残基K154/155被证明是体内致癌ErbB2驱动的乳腺癌生长和乳腺肿瘤干细胞扩增所必需的。调控生长因子信号、脂肪生成、细胞凋亡和干细胞扩增的转录模块的诱导依赖于K154/155。K154/155残基的乙酰化状态决定了全基因组DNA结合位点的选择,改变了从规范到非规范(C/EBP) DNA序列特异性结合的选择。反映脂肪生成中乙酰化依赖性基因组占用的基因特征对ErbB2+乳腺癌的生存结果具有预测价值。parar γ - 1乙酰化位点对erbb2诱导的乳腺癌肿瘤生长至关重要,可能是治疗共灭的相关靶点。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Oncogene
医学-生化与分子生物学
CiteScore
15.30
自引率
1.20%
发文量
404
审稿时长
1 months
期刊介绍:
Oncogene is dedicated to advancing our understanding of cancer processes through the publication of exceptional research. The journal seeks to disseminate work that challenges conventional theories and contributes to establishing new paradigms in the etio-pathogenesis, diagnosis, treatment, or prevention of cancers. Emphasis is placed on research shedding light on processes driving metastatic spread and providing crucial insights into cancer biology beyond existing knowledge.
Areas covered include the cellular and molecular biology of cancer, resistance to cancer therapies, and the development of improved approaches to enhance survival. Oncogene spans the spectrum of cancer biology, from fundamental and theoretical work to translational, applied, and clinical research, including early and late Phase clinical trials, particularly those with biologic and translational endpoints.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


