Multi-scale interaction and locally enhanced bridging network for medical image segmentation
IF 4.9
2区 医学
Q1 ENGINEERING, BIOMEDICAL
Computerized Medical Imaging and Graphics
Pub Date : 2025-07-22
DOI:10.1016/j.compmedimag.2025.102610
引用次数: 0
Abstract
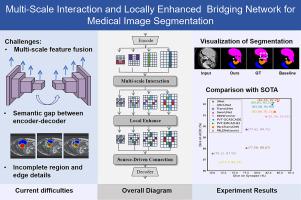
Accurate organ segmentation is crucial for precise medical diagnosis. Recent methods in CNNs and Transformers have significantly enhanced automatic medical image segmentation. Their encoders and decoders often rely on simple skip connections, which fail to effectively integrate multi-scale features. This causes a misalignment between low-resolution global features and high-resolution spatial information. As a result, segmentation accuracy suffers, particularly in global contours and local details. To address this limitation, MILENet, a multi-scale interaction and locally enhanced bridging network, is proposed. The proposed context bridge incorporates a multi-scale interaction module to reorganize multi-scale features and ensure global correlation. Additionally, a local enhancement module is introduced. It includes a dilated coordinate attention mechanism and a locally enhanced FFN built with a cascaded convolutional structure. This module enhances local context modeling and improves feature discrimination. Furthermore, a source-driven connection mechanism is introduced to preserve detailed information across layers, providing richer features for decoder reconstruction. By leveraging these innovations, MILENet effectively aligns multi-scale features and enhances local details, thereby improving segmentation accuracy. MILENet has been evaluated on publicly available datasets spanning abdominal CT (Synapse), cardiac MRI (ACDC), and colonoscopy RGB images (Kvasir, CVC-ClinicDB, CVC-ColonDB, CVC-300, and ETIS-LaribDB). The results show that MILENet achieves state-of-the-art performance across different modalities. It effectively handles both large-organ segmentation in CT/MRI and fine-grained polyp delineation in endoscopic images, demonstrating strong generalizability to diverse anatomical structures and imaging conditions. The code has been released on GitHub: https://github.com/syzhou1226/MILENET.
基于多尺度交互和局部增强桥接网络的医学图像分割
准确的器官分割对于精确的医学诊断至关重要。最近的cnn和transformer方法显著增强了医学图像的自动分割。它们的编码器和解码器往往依赖于简单的跳过连接,无法有效地集成多尺度特征。这导致了低分辨率全局特征和高分辨率空间信息之间的不对齐。因此,分割精度受到影响,特别是在全局轮廓和局部细节方面。为了解决这一限制,提出了一种多尺度交互和局部增强桥接网络MILENet。本文提出的上下文桥包含一个多尺度交互模块,以重组多尺度特征并确保全局关联。此外,还引入了局部增强模块。它包括一个扩展的坐标注意机制和一个用级联卷积结构构建的局部增强FFN。该模块增强了局部上下文建模,改进了特征识别。此外,引入了源驱动的连接机制,以跨层保存详细信息,为解码器重建提供更丰富的功能。通过利用这些创新,MILENet有效地对齐多尺度特征并增强局部细节,从而提高分割精度。MILENet已在公开可用的数据集上进行了评估,这些数据集包括腹部CT (Synapse)、心脏MRI (ACDC)和结肠镜RGB图像(Kvasir、CVC-ClinicDB、CVC-ColonDB、CVC-300和ETIS-LaribDB)。结果表明,MILENet在不同模态下实现了最先进的性能。它能有效地处理CT/MRI中的大器官分割和内镜图像中细粒度息肉的描绘,对不同的解剖结构和成像条件具有很强的通用性。代码已经在GitHub上发布:https://github.com/syzhou1226/MILENET。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
CiteScore
10.70
自引率
3.50%
发文量
71
审稿时长
26 days
期刊介绍:
The purpose of the journal Computerized Medical Imaging and Graphics is to act as a source for the exchange of research results concerning algorithmic advances, development, and application of digital imaging in disease detection, diagnosis, intervention, prevention, precision medicine, and population health. Included in the journal will be articles on novel computerized imaging or visualization techniques, including artificial intelligence and machine learning, augmented reality for surgical planning and guidance, big biomedical data visualization, computer-aided diagnosis, computerized-robotic surgery, image-guided therapy, imaging scanning and reconstruction, mobile and tele-imaging, radiomics, and imaging integration and modeling with other information relevant to digital health. The types of biomedical imaging include: magnetic resonance, computed tomography, ultrasound, nuclear medicine, X-ray, microwave, optical and multi-photon microscopy, video and sensory imaging, and the convergence of biomedical images with other non-imaging datasets.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


