Machine learning reveals complex genetics of fungal resistance in sorghum grain mold
IF 3.9
2区 生物学
Q2 ECOLOGY
引用次数: 0
Abstract
Plant disease resistance is often a complex, polygenic trait, making its genetic dissection with traditional genome-wide association studies (GWAS) challenging. Grain mold in sorghum, a devastating disease caused by a fungal complex, exemplifies this complexity. We hypothesized that a machine learning (ML)-driven GWAS, employing diverse phenotypic representations from a panel of 306 sorghum accessions, could more effectively unravel the genetic basis of resistance. Phenotypic data, including raw disease scores, a ‘difference phenotype’ (inoculated vs. control), and principal components, were analyzed using Boosted Tree and Bootstrap Forest models, demonstrating strong explanatory power for phenotypic variance when trained on the entire dataset. This ML-GWAS approach confirmed a highly polygenic architecture for grain mold resistance, identifying numerous SNPs across the sorghum genome. Notably, several SNPs were consistently associated with resistance across multiple analytical models and phenotypic representations. These robustly identified SNPs were frequently located near genes with predicted functions integral to plant defense. Gene ontology (GO) analyses of the candidate gene set confirmed enrichment in categories supporting roles in pathogen recognition, DNA repair, and stress response modulation, indicating a multifaceted defense mechanism. This study provides valuable candidate genes for breeding sorghum with enhanced grain mold resistance and offers a refined methodological framework for dissecting complex traits in this crop. The successful application of this ML-based strategy in sorghum suggests its potential utility for studying similar complex traits in other plant species.
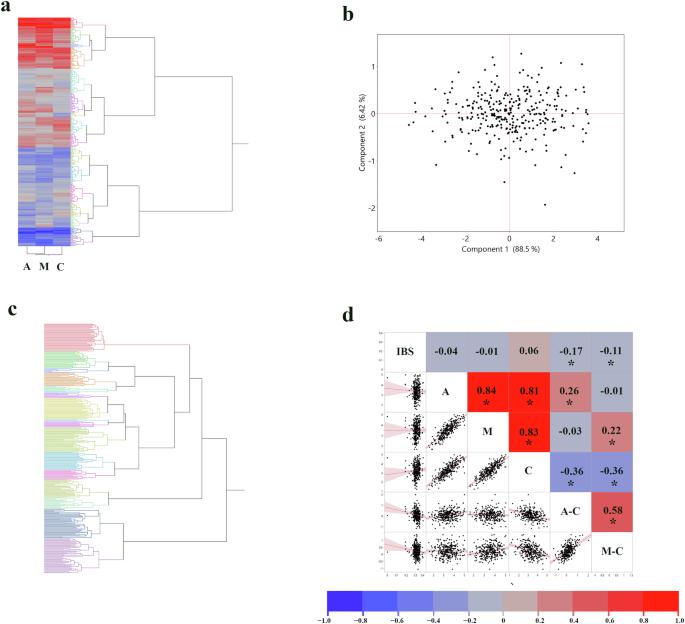
机器学习揭示了高粱谷物霉菌抗性的复杂遗传。
植物抗病性通常是一个复杂的多基因性状,这使得传统的全基因组关联研究(GWAS)对其进行遗传解剖具有挑战性。高粱的谷粒霉是一种由真菌复合体引起的毁灭性疾病,它体现了这种复杂性。我们假设,采用306份高粱材料的不同表型表征的机器学习驱动的GWAS可以更有效地揭示抗性的遗传基础。表型数据,包括原始疾病评分、“差异表型”(接种与对照)和主成分,使用boost Tree和Bootstrap Forest模型进行分析,在整个数据集上进行训练时,显示出对表型方差的强大解释力。这种ML-GWAS方法证实了谷物抗霉菌性的高多基因结构,鉴定了高粱基因组中的许多snp。值得注意的是,几个snp在多个分析模型和表型表征中始终与抗性相关。这些已确定的snp通常位于与植物防御相关的预测功能基因附近。对候选基因集的基因本体(GO)分析证实了在病原体识别、DNA修复和应激反应调节中起支持作用的类别的富集,表明其具有多方面的防御机制。本研究为选育具有较强抗霉性的高粱提供了有价值的候选基因,并为剖析该作物的复杂性状提供了完善的方法框架。这种基于ml的策略在高粱上的成功应用表明,它在研究其他植物物种的类似复杂性状方面具有潜在的实用性。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Heredity
生物-进化生物学
CiteScore
7.50
自引率
2.60%
发文量
84
审稿时长
4-8 weeks
期刊介绍:
Heredity is the official journal of the Genetics Society. It covers a broad range of topics within the field of genetics and therefore papers must address conceptual or applied issues of interest to the journal''s wide readership
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


