Integrated multidimensional bioinformatics analysis of the molecular mechanisms of ulcerative colitis-associated colorectal cancer and MMP1 as a potential therapeutic target
IF 5
3区 医学
Q1 BIOTECHNOLOGY & APPLIED MICROBIOLOGY
引用次数: 0
Abstract
This study aimed to investigate the molecular mechanisms underlying ulcerative colitis (UC)-associated colorectal cancer (CRC) development and identify potential therapeutic targets through integrated multi-omics analysis. Mendelian randomization (MR) analysis, combined with bioinformatics approaches including differential gene expression analysis, protein-protein interaction network construction, gene set enrichment analysis, and single-cell RNA sequencing, was employed. Data were obtained from GEO, TCGA, and genome-wide association study (GWAS) databases. Drug prediction and molecular docking were performed using DSigDB and AutoDockTools. A total of 48 shared genes were identified between UC and CRC, with MMP1 emerging as a significant protective factor (OR = 0.766; 95% CI = 0.593–0.989, P = 0.041). MMP1 demonstrated strong diagnostic potential (AUC = 0.927, 95% CI = 0.895–0.959) and was functionally associated with immune regulation and metabolic pathways. Single-cell analysis revealed predominant MMP1 expression in fibroblasts and immune cells, while immune infiltration analysis showed significant correlations with CD8⁺ T cells and NK cells. Mediation MR analysis indicated that 63.33% of MMP1’s protective effect was mediated through naive-mature B cells. Drug prediction identified ilomastat as a potential MMP1 inhibitor with strong binding affinity (binding energy = –7.17 kcal/mol). These findings provide evidence for MMP1’s protective role in UC-associated CRC through immune microenvironment modulation, highlighting its potential as a diagnostic biomarker and therapeutic target. The identification of ilomastat as a potential MMP1 inhibitor offers new avenues for targeted therapy in inflammation-associated cancers.
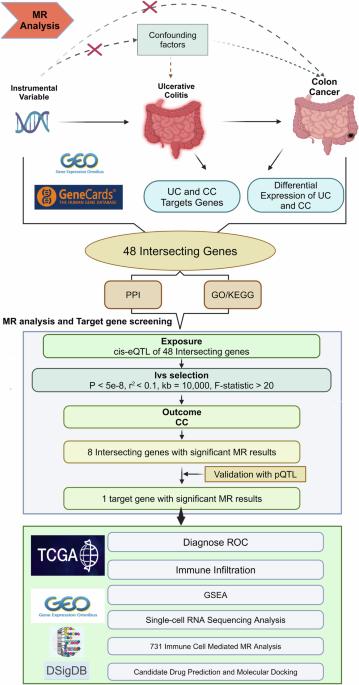
综合多维生物信息学分析溃疡性结肠炎相关结直肠癌的分子机制和MMP1作为潜在的治疗靶点。
本研究旨在通过综合多组学分析探讨溃疡性结肠炎(UC)相关结直肠癌(CRC)发展的分子机制,并确定潜在的治疗靶点。采用孟德尔随机化(MR)分析,结合生物信息学方法,包括差异基因表达分析、蛋白质相互作用网络构建、基因集富集分析和单细胞RNA测序。数据来自GEO、TCGA和全基因组关联研究(GWAS)数据库。使用DSigDB和AutoDockTools进行药物预测和分子对接。UC和CRC共有48个共享基因,其中MMP1是一个重要的保护因子(OR = 0.766;95% ci = 0.593-0.989, p = 0.041)。MMP1表现出较强的诊断潜力(AUC = 0.927, 95% CI = 0.895-0.959),并与免疫调节和代谢途径相关。单细胞分析显示MMP1在成纤维细胞和免疫细胞中主要表达,免疫浸润分析显示CD8 +与T细胞和NK细胞有显著相关性。介导MR分析表明,63.33%的MMP1保护作用是通过幼稚成熟B细胞介导的。药物预测发现伊洛司他是一种潜在的MMP1抑制剂,具有很强的结合亲和力(结合能= -7.17 kcal/mol)。这些发现为MMP1通过免疫微环境调节在uc相关结直肠癌中发挥保护作用提供了证据,突出了其作为诊断生物标志物和治疗靶点的潜力。伊洛马司他作为潜在的MMP1抑制剂的鉴定为炎症相关癌症的靶向治疗提供了新的途径。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Cancer gene therapy
医学-生物工程与应用微生物
CiteScore
10.20
自引率
0.00%
发文量
150
审稿时长
4-8 weeks
期刊介绍:
Cancer Gene Therapy is the essential gene and cellular therapy resource for cancer researchers and clinicians, keeping readers up to date with the latest developments in gene and cellular therapies for cancer. The journal publishes original laboratory and clinical research papers, case reports and review articles. Publication topics include RNAi approaches, drug resistance, hematopoietic progenitor cell gene transfer, cancer stem cells, cellular therapies, homologous recombination, ribozyme technology, antisense technology, tumor immunotherapy and tumor suppressors, translational research, cancer therapy, gene delivery systems (viral and non-viral), anti-gene therapy (antisense, siRNA & ribozymes), apoptosis; mechanisms and therapies, vaccine development, immunology and immunotherapy, DNA synthesis and repair.
Cancer Gene Therapy publishes the results of laboratory investigations, preclinical studies, and clinical trials in the field of gene transfer/gene therapy and cellular therapies as applied to cancer research. Types of articles published include original research articles; case reports; brief communications; review articles in the main fields of drug resistance/sensitivity, gene therapy, cellular therapy, tumor suppressor and anti-oncogene therapy, cytokine/tumor immunotherapy, etc.; industry perspectives; and letters to the editor.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


