Identifying homogenous patient subgroups using transformer based hierarchical clustering of heterogeneous Mixed-Modality medical data
IF 4.5
2区 医学
Q2 COMPUTER SCIENCE, INTERDISCIPLINARY APPLICATIONS
引用次数: 0
Abstract
Objective
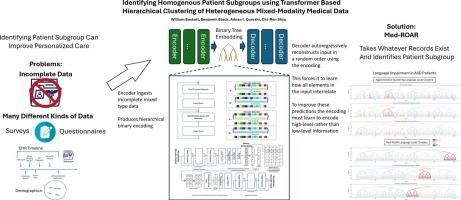
Patients are highly heterogeneous, with varying needs and responses to treatment. Identifying clinically homogenous patient subgroups is critical to improve personalized care. Patient records are often heterogeneous, may include multiple modalities which conventionally require separate data processing considerations, and are often incomplete, leading to difficulties in identifying meaningful clusters of patients.
Methods
We introduce a Med-ROAR, a transformer-based Random Order AutoRegressive (ROAR) embedding model for medical data. Med-ROAR hierarchically clusters data by encoding it into hierarchical discrete embeddings using a modified self-attention operation to facilitate random order mixed modality autoregressive modeling. This allows the model to accept arbitrary mixes of record types without special considerations. We compare our method’s clustering effectiveness to standard agglomerative clustering using 147,469 individuals diagnosed with Autism Spectrum Disorder (ASD). We also evaluate its use on data with mixed modalities and its resilience to missing information using 50,458 clinical records from Intensive Care Unit (ICU) patients which include both tabular and time-series components.
Results
We demonstrate that Med-ROAR is more likely to discover more cohesive high-level clusters than distance-based methods like agglomerative clustering. Our exploratory analysis of the autism data identifies clinically meaningful patterns of phenotypes within ASD. We identify homogenous, but atypical, patient subgroups within the ASD population. We also demonstrate Med-ROAR’s effectiveness in clustering patients using mixes of both tabular and time series clinical records from ICU patients. We demonstrate that Med-ROAR can predict patient subgroups even using incomplete, preliminary information collected shortly after admission.
Conclusion
Med-ROAR is a flexible hierarchical clustering technique which learns to cluster patients based on learned high-level semantic similarities rather than rule-based metrics. It can accept whatever patient data may be available without modification to the underlying model architecture. The data modalities which Med-ROAR can accept are primarily constrained by computational resources, rather than architectural limitations.
使用基于变压器的异构混合模式医疗数据分层聚类识别同质患者亚组。
目的:患者具有高度异质性,对治疗的需求和反应各不相同。确定临床同质患者亚组对于改善个性化护理至关重要。患者记录通常是异构的,可能包括多种模式,通常需要单独的数据处理考虑,并且通常是不完整的,导致难以识别有意义的患者群。方法:引入一种基于变压器的医疗数据随机顺序自回归嵌入模型Med-ROAR。Med-ROAR通过使用改进的自注意操作将数据编码成分层离散嵌入来分层聚类数据,以促进随机顺序混合模态自回归建模。这允许模型接受记录类型的任意混合,而无需特别考虑。我们使用147,469名自闭症谱系障碍(ASD)患者,将我们的方法与标准聚集聚类的聚类效果进行了比较。我们还利用重症监护病房(ICU)患者的50,458份临床记录(包括表格和时间序列成分)评估了其对混合模式数据的使用及其对缺失信息的恢复能力。结果:我们证明Med-ROAR比基于距离的方法(如聚集聚类)更有可能发现更具内聚性的高层聚类。我们对自闭症数据的探索性分析确定了ASD中具有临床意义的表型模式。我们在ASD人群中识别同质但非典型的患者亚群。我们还证明了Med-ROAR在使用来自ICU患者的表格和时间序列临床记录混合聚类患者方面的有效性。我们证明,即使使用入院后不久收集的不完整的初步信息,Med-ROAR也可以预测患者亚组。结论:Med-ROAR是一种灵活的分层聚类技术,它基于学习到的高级语义相似性而不是基于规则的度量来学习聚类患者。它可以接受任何可用的患者数据,而无需修改底层模型体系结构。Med-ROAR可以接受的数据模式主要受到计算资源的限制,而不是架构的限制。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Biomedical Informatics
医学-计算机:跨学科应用
CiteScore
8.90
自引率
6.70%
发文量
243
审稿时长
32 days
期刊介绍:
The Journal of Biomedical Informatics reflects a commitment to high-quality original research papers, reviews, and commentaries in the area of biomedical informatics methodology. Although we publish articles motivated by applications in the biomedical sciences (for example, clinical medicine, health care, population health, and translational bioinformatics), the journal emphasizes reports of new methodologies and techniques that have general applicability and that form the basis for the evolving science of biomedical informatics. Articles on medical devices; evaluations of implemented systems (including clinical trials of information technologies); or papers that provide insight into a biological process, a specific disease, or treatment options would generally be more suitable for publication in other venues. Papers on applications of signal processing and image analysis are often more suitable for biomedical engineering journals or other informatics journals, although we do publish papers that emphasize the information management and knowledge representation/modeling issues that arise in the storage and use of biological signals and images. System descriptions are welcome if they illustrate and substantiate the underlying methodology that is the principal focus of the report and an effort is made to address the generalizability and/or range of application of that methodology. Note also that, given the international nature of JBI, papers that deal with specific languages other than English, or with country-specific health systems or approaches, are acceptable for JBI only if they offer generalizable lessons that are relevant to the broad JBI readership, regardless of their country, language, culture, or health system.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


