Time resolved cell painting enables rapid assessment of cell phenotypes
IF 2.7
4区 生物学
Q2 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
Abstract
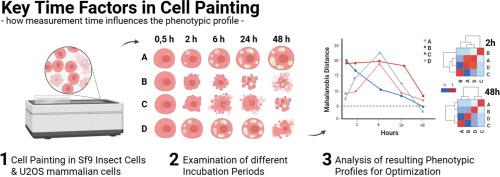
Cell Painting is an advanced imaging technique for drug discovery used to study cellular phenotypes by simultaneously labeling various organelles/structures and analyzing the resulting multidimensional phenotypic features through a sophisticated data analysis pipeline. Based on established phenotyping methodologies, this method has relied on incubation times of typically around 48 h for the assessment of phenotypic fingerprints. Here we provide evidence that earlier assessments show more robust results with increased significance of phenotypic fingerprints that better reflect primary physiological effects.
Our study included compounds that range from representatives with modes of action that result in immediate phenotypic changes, such as energy metabolism inhibitors, to representatives that typically show pronounced phenotypes after several days, such as developmental inhibitors. Remarkably, we observed that for all compounds, primary cellular alterations were best detected at early timepoints after treatment, specifically at 6 h for Sf9 insect cells and shortly later timepoints for mammalian U2OS cells. Brief incubation periods enable the capture of primary effects of treatments while minimizing the influence of secondary changes as well as downstream phenotypic alterations like, for example, cell death. This enhances the specificity and accuracy of Cell Painting and consequently provides a more immediate depiction of primary actions from compounds. Notably, it also improves the efficiency of experimental workflows.
In conclusion, we propose a more rapid assessment of cell phenotypes and morphology in the Cell Painting assay to enable a higher throughput in drug discovery screenings.
时间分辨细胞绘画能够快速评估细胞表型。
细胞绘画是一种先进的成像技术,用于药物发现,通过同时标记各种细胞器/结构,并通过复杂的数据分析管道分析由此产生的多维表型特征,用于研究细胞表型。基于已建立的表型方法,该方法依赖于典型的48小时孵育时间来评估表型指纹。在这里,我们提供的证据表明,早期的评估显示了更稳健的读数,表型指纹的重要性增加,更好地反映了主要的生理效应。我们的研究包括从具有直接导致表型变化的作用模式的代表化合物,如能量代谢抑制剂,到通常在几天后显示显着表型的代表化合物,如发育抑制剂。值得注意的是,我们观察到,对于所有化合物,在处理后的早期时间点,尤其是在Sf9昆虫细胞的6小时和哺乳动物U2OS细胞的较晚时间点,可以最好地检测到细胞的原代改变。短暂的潜伏期能够捕获治疗的主要效果,并最大限度地减少继发性变化的影响以及下游表型改变,例如细胞死亡。这提高了细胞绘画的特异性和准确性,从而提供了化合物的主要作用的更直接的描述。值得注意的是,它还提高了实验工作流程的效率。总之,我们建议在细胞涂色试验中更快速地评估细胞表型和形态,以提高药物发现筛选的通量。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

SLAS Discovery
Chemistry-Analytical Chemistry
CiteScore
7.00
自引率
3.20%
发文量
58
审稿时长
39 days
期刊介绍:
Advancing Life Sciences R&D: SLAS Discovery reports how scientists develop and utilize novel technologies and/or approaches to provide and characterize chemical and biological tools to understand and treat human disease.
SLAS Discovery is a peer-reviewed journal that publishes scientific reports that enable and improve target validation, evaluate current drug discovery technologies, provide novel research tools, and incorporate research approaches that enhance depth of knowledge and drug discovery success.
SLAS Discovery emphasizes scientific and technical advances in target identification/validation (including chemical probes, RNA silencing, gene editing technologies); biomarker discovery; assay development; virtual, medium- or high-throughput screening (biochemical and biological, biophysical, phenotypic, toxicological, ADME); lead generation/optimization; chemical biology; and informatics (data analysis, image analysis, statistics, bio- and chemo-informatics). Review articles on target biology, new paradigms in drug discovery and advances in drug discovery technologies.
SLAS Discovery is of particular interest to those involved in analytical chemistry, applied microbiology, automation, biochemistry, bioengineering, biomedical optics, biotechnology, bioinformatics, cell biology, DNA science and technology, genetics, information technology, medicinal chemistry, molecular biology, natural products chemistry, organic chemistry, pharmacology, spectroscopy, and toxicology.
SLAS Discovery is a member of the Committee on Publication Ethics (COPE) and was published previously (1996-2016) as the Journal of Biomolecular Screening (JBS).
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


