Self-iterative multiple-instance learning enables the prediction of CD4+ T cell immunogenic epitopes
IF 23.9
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
Abstract
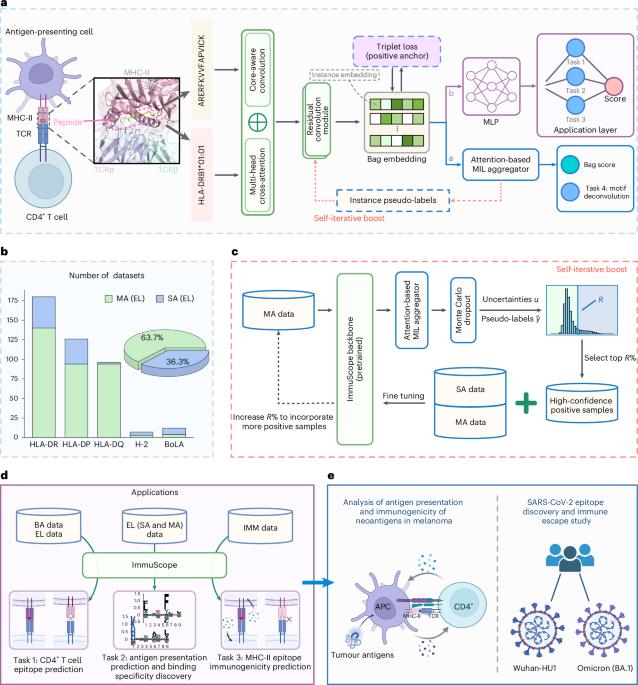
Accurate prediction of antigen presentation to CD4+ T cells and subsequent induction of immune response are fundamentally important for vaccine development, autoimmune disease treatment and cancer neoepitope discovery. In immunopeptidomics, single-allelic data offer high specificity but limited allele coverage, whereas multi-allelic data provide broader representation at the expense of weak labelling. Current computational approaches either overlook the abundance of multi-allelic data or suffer from label ambiguity due to inadequate modelling strategies. To address these limitations, we present ImmuScope, a weakly supervised deep learning framework that integrates major histocompatibility complex class II (MHC-II) antigen presentation, CD4+ T cell epitopes and immunogenicity assessment. ImmuScope leverages self-iterative multiple-instance learning with positive-anchor triplet loss to decipher peptide-MHC-II binding from weakly labelled multi-allelic data and high-confidence single-allelic data. The training dataset comprises over 600,000 ligands across 142 alleles. Additionally, ImmuScope enables the interpretation of MHC-II binding specificity and motif deconvolution of immunopeptidomics data. We successfully applied ImmuScope to identify melanoma neoantigens, uncovering mutation-driven variations in peptide-MHC-II binding and immunogenicity. Furthermore, we employed ImmuScope to evaluate the effects of SARS-CoV-2 epitope mutations associated with immune escape, with predictions well aligned with experimentally observed immune escape dynamics. Overall, by offering a unified solution for CD4+ T cell antigen recognition and immunogenicity assessment, ImmuScope holds substantial promise for accelerating vaccine design and advancing personalized immunotherapy. ImmuScope, a weakly supervised deep learning model capable of analysing multi- and single-allelic data, is introduced, facilitating interpretable neoantigen discovery and immune escape analysis.

自迭代多实例学习能够预测CD4+ T细胞免疫原性表位
准确预测抗原呈递到CD4+ T细胞并随后诱导免疫应答对于疫苗开发、自身免疫性疾病治疗和癌症新表位发现至关重要。在免疫肽组学中,单等位基因数据提供了高特异性但有限的等位基因覆盖,而多等位基因数据以弱标记为代价提供了更广泛的代表性。目前的计算方法要么忽略了多等位基因数据的丰富性,要么由于建模策略不充分而导致标签模糊。为了解决这些限制,我们提出了ImmuScope,这是一个弱监督深度学习框架,集成了主要组织相容性复合体II类(MHC-II)抗原呈递、CD4+ T细胞表位和免疫原性评估。ImmuScope利用自迭代多实例学习和正锚三重态丢失,从弱标记的多等位基因数据和高置信度的单等位基因数据中破译肽- mhc - ii结合。训练数据集包含142个等位基因,超过60万个配体。此外,ImmuScope能够解释MHC-II结合特异性和免疫肽组学数据的基序反褶积。我们成功地应用ImmuScope识别黑色素瘤新抗原,揭示了肽- mhc - ii结合和免疫原性的突变驱动变异。此外,我们利用ImmuScope评估了SARS-CoV-2表位突变与免疫逃逸相关的影响,预测结果与实验观察到的免疫逃逸动力学非常吻合。总的来说,通过提供CD4+ T细胞抗原识别和免疫原性评估的统一解决方案,ImmuScope在加速疫苗设计和推进个性化免疫治疗方面具有重大前景。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Machine Intelligence
Multiple-
CiteScore
36.90
自引率
2.10%
发文量
127
期刊介绍:
Nature Machine Intelligence is a distinguished publication that presents original research and reviews on various topics in machine learning, robotics, and AI. Our focus extends beyond these fields, exploring their profound impact on other scientific disciplines, as well as societal and industrial aspects. We recognize limitless possibilities wherein machine intelligence can augment human capabilities and knowledge in domains like scientific exploration, healthcare, medical diagnostics, and the creation of safe and sustainable cities, transportation, and agriculture. Simultaneously, we acknowledge the emergence of ethical, social, and legal concerns due to the rapid pace of advancements.
To foster interdisciplinary discussions on these far-reaching implications, Nature Machine Intelligence serves as a platform for dialogue facilitated through Comments, News Features, News & Views articles, and Correspondence. Our goal is to encourage a comprehensive examination of these subjects.
Similar to all Nature-branded journals, Nature Machine Intelligence operates under the guidance of a team of skilled editors. We adhere to a fair and rigorous peer-review process, ensuring high standards of copy-editing and production, swift publication, and editorial independence.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


