Exploring Complexity-Calibrated morphological distribution for whole slide image classification and difficulty-grading
IF 11.8
1区 医学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
Abstract
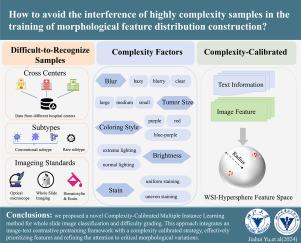
Multiple Instance Learning (MIL) is essential for accurate pathological image classification under limited annotations. Global-local morphological modeling approaches have shown promise in whole slide image (WSI) analysis by aligning patches with spatial positions. However, these methods fail to differentiate samples by complexity during morphological distribution construction, treating all samples equally important for model training. This oversight disregards the impact of difficult-to-recognize samples, leading to a morphological fitting bottleneck that hinders the clinical application of deep learning across centers, subtypes, and imaging standards. To address this, we propose Complexity-Calibrated MIL (CoCaMIL) for WSI classification and difficulty grading. CoCaMIL emphasizes the synergistic effects between morphological distribution and key complexity factors, including blur, tumor size, coloring style, brightness, and stain. Specifically, we developed an image–text contrastive pretraining framework to jointly learn multiple complexity factors, enhancing morphological distribution fitting. Additionally, to reduce the tendency to focus on difficult samples overly, we introduce a complexity calibration method, which forms a distance-prioritized feature distribution by incorporating objective factors during training. CoCaMIL achieved top classification performance across three large benchmarks and established a reliable system for grading sample difficulty. To our knowledge, CoCaMIL is the first approach to construct WSI morphological representations based on the collaborative integration of complexity factors, offering a new perspective to broaden the clinical use of deep learning in digital pathology. The code is available at https://github.com/sm8754/cocamil.
探索基于复杂度校正的形态学分布在全幻灯片图像分类和难度分级中的应用
多实例学习(MIL)是在有限注释下对病理图像进行准确分类的关键。全局-局部形态学建模方法在整个幻灯片图像(WSI)分析中显示出前景,该方法通过对齐斑块与空间位置。然而,这些方法在形态分布构建过程中无法通过复杂性来区分样本,在模型训练中对所有样本都同等重要。这种疏忽忽视了难以识别的样本的影响,导致形态拟合瓶颈,阻碍了跨中心、亚型和成像标准的深度学习的临床应用。为了解决这个问题,我们提出了用于WSI分类和难度分级的复杂性校准MIL (CoCaMIL)。CoCaMIL强调形态学分布与关键复杂性因素之间的协同作用,包括模糊、肿瘤大小、着色风格、亮度和染色。具体而言,我们开发了一个图像-文本对比预训练框架,共同学习多个复杂性因素,增强形态分布拟合。此外,为了减少过度关注困难样本的倾向,我们引入了一种复杂性校准方法,该方法通过在训练过程中结合客观因素形成距离优先的特征分布。CoCaMIL在三个大型基准中取得了最高的分类性能,并建立了可靠的样本难度分级系统。据我们所知,CoCaMIL是第一个基于复杂性因素协同集成构建WSI形态学表征的方法,为拓宽深度学习在数字病理学中的临床应用提供了新的视角。代码可在https://github.com/sm8754/cocamil上获得。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Medical image analysis
工程技术-工程:生物医学
CiteScore
22.10
自引率
6.40%
发文量
309
审稿时长
6.6 months
期刊介绍:
Medical Image Analysis serves as a platform for sharing new research findings in the realm of medical and biological image analysis, with a focus on applications of computer vision, virtual reality, and robotics to biomedical imaging challenges. The journal prioritizes the publication of high-quality, original papers contributing to the fundamental science of processing, analyzing, and utilizing medical and biological images. It welcomes approaches utilizing biomedical image datasets across all spatial scales, from molecular/cellular imaging to tissue/organ imaging.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


