Unbalanced gene-level batch effects in single-cell data
IF 18.3
Q1 COMPUTER SCIENCE, INTERDISCIPLINARY APPLICATIONS
引用次数: 0
Abstract
We developed group technical effects (GTE) as a quantitative metric for evaluating gene-level batch effects in single-cell data. It identifies highly batch-sensitive genes — the primary contributors to batch effects — that vary across datasets, and whose removal effectively mitigates the batch effects.
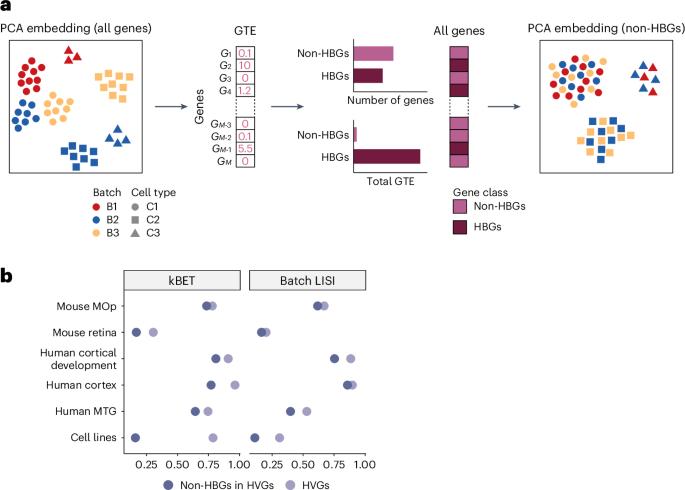
单细胞数据中不平衡基因水平的批效应。
我们开发了群体技术效应(GTE)作为评估单细胞数据中基因水平批效应的定量指标。它确定了高度批次敏感的基因——批次效应的主要贡献者——这些基因在不同的数据集上有所不同,删除它们可以有效地减轻批次效应。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


