Iterative variational learning of committor-consistent transition pathways using artificial neural networks
IF 18.3
Q1 COMPUTER SCIENCE, INTERDISCIPLINARY APPLICATIONS
引用次数: 0
Abstract
Discovering transition pathways that are physically meaningful and committor-consistent has long been a challenge in studying rare events in complex systems. Here we introduce a neural network-based strategy that learns simultaneously the committor function and the associated committor-consistent string, offering an unprecedented view of transition processes. Built on the committor time-correlation function, this method operates across diverse dynamical regimes, and extends beyond traditional approaches relying on infinitesimal time-lag approximations, valid only in the overdamped diffusive limit. It also distinguishes multiple competing pathways, crucial for understanding complex biomolecular transformations. Demonstrated on benchmark potentials and biological systems such as peptide isomerization and protein-model folding, this approach robustly reproduces established dynamics, rate constants and transition mechanisms. Its adaptability to collective variables and resilience across neural architectures make it a powerful and versatile tool for enhanced-sampling simulations of rare events, enabling insights into the intricate landscapes of biomolecular systems. A neural network approach grounded in transition-path theory is shown to uncover committor-consistent transition pathways, resolving competing mechanisms across dynamical regimes and advancing the modeling of rare events in biomolecular systems.
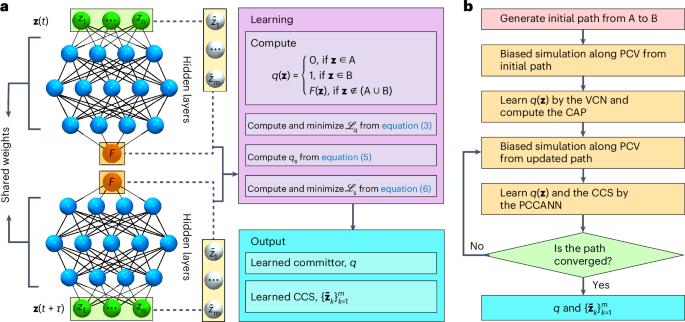
基于人工神经网络的提交者一致转换路径的迭代变分学习。
长期以来,在研究复杂系统中的罕见事件时,发现物理上有意义且与提交者一致的转换路径一直是一个挑战。在这里,我们介绍了一种基于神经网络的策略,该策略同时学习提交者函数和相关的提交者一致字符串,提供了一个前所未有的过渡过程视图。该方法建立在提交者时间相关函数的基础上,可以在不同的动态状态下运行,并且超越了依赖于仅在过阻尼扩散极限下有效的无穷小时滞近似的传统方法。它还区分了多种竞争途径,这对于理解复杂的生物分子转化至关重要。在基准电位和生物系统(如肽异构化和蛋白质模型折叠)上进行了验证,该方法可靠地再现了已建立的动力学、速率常数和过渡机制。它对集体变量的适应性和跨神经结构的弹性使其成为一种强大而通用的工具,用于增强罕见事件的采样模拟,使人们能够深入了解生物分子系统的复杂景观。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


