RNA–ligand interaction scoring via data perturbation and augmentation modeling
IF 18.3
Q1 COMPUTER SCIENCE, INTERDISCIPLINARY APPLICATIONS
引用次数: 0
Abstract
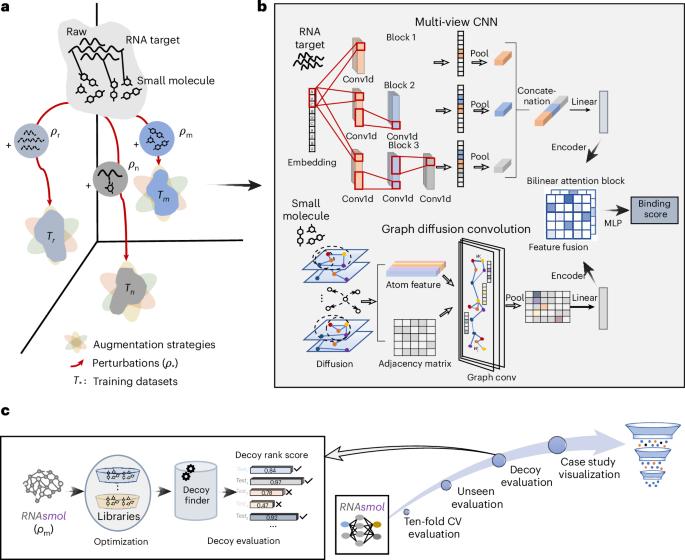
Despite recent advances in RNA-targeting drug discovery, the development of data-driven deep learning models remains challenging owing to limited validated RNA–small molecule interaction data and scarce known RNA structures. In this context, we introduce RNAsmol, a sequence-based deep learning framework that incorporates data perturbation with augmentation, graph-based molecular feature representation and attention-based feature fusion modules to predict RNA–small molecule interactions. RNAsmol employs perturbation strategies to balance the bias between the true negative and unknown interaction space, thereby elucidating the intrinsic binding patterns between RNA and small molecules. The resulting model demonstrates accurate predictions of the binding between RNA and small molecules, outperforming other methods in ten-fold cross-validation, unseen evaluation and decoy evaluation. Moreover, we use case studies to visualize molecular binding profiles and the distribution of learned weights, providing interpretable insights into RNAsmol’s predictions. In particular, without requiring structural input, RNAsmol can generate reliable predictions and be adapted to various drug design scenarios. RNAsmol is a sequence-based deep learning framework for predicting RNA–small molecule interactions, integrating perturbation and augmentation to achieve robust predictions under limited data and adaptability to diverse drug discovery scenarios.
通过数据扰动和增强模型进行rna -配体相互作用评分。
尽管最近在RNA靶向药物发现方面取得了进展,但由于有效的RNA-小分子相互作用数据有限,已知的RNA结构很少,数据驱动的深度学习模型的开发仍然具有挑战性。在这种背景下,我们引入了RNAsmol,一个基于序列的深度学习框架,它结合了数据扰动和增强,基于图的分子特征表示和基于注意力的特征融合模块来预测rna -小分子相互作用。RNAsmol采用微扰策略来平衡真负相互作用空间和未知相互作用空间之间的偏差,从而阐明RNA与小分子之间的内在结合模式。所得到的模型能够准确预测RNA和小分子之间的结合,在十倍交叉验证、未见评估和诱饵评估方面优于其他方法。此外,我们使用案例研究来可视化分子结合概况和学习权的分布,为RNAsmol的预测提供可解释的见解。特别是,在不需要结构输入的情况下,RNAsmol可以产生可靠的预测,并适应各种药物设计场景。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


