Identification of 1-O-Galloyl -β-D-glucose as a potent activator of Sirtuin-1: an in-silico study
IF 3
4区 生物学
Q2 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
Abstract
Sirt-1 is a deacetylase acting on histones and various non-histone proteins, playing a crucial role in multiple physiological and pathological processes. Scientific efforts to regulate its activity primarily focus on small molecule activators. In order to identify better small molecular natural compounds activating Sirt-1, this study employed traditional knowledge-driven in silico studies based on phytochemicals from selected medicinal plants. Molecular docking studies against Sirt-1 using a phytochemical library, identified 1-O-galloyl-β-D-Glucose (GBDG) that binds to the allosteric site of Sirt-1 with docking score, H-bond interaction and other docking features better than that of resveratrol, a known natural small molecular activator of Sirt-1. Molecular dynamic simulation of the docked complex, followed by trajectory analysis (RMSD, RMSF, Radius of Gyration and binding energy) demonstrated that the complex is structurally and thermodynamically stable. Centroid distance measurement between key residues in the regulatory and catalytic domain revealed that docking of GBDG resulted in change in conformation fetching catalytic domain closer to the regulatory domain. GBDG docking against Sirt-1 increased its affinity to the acetylated substrate as indicated by better docking parameters when compared with that of Sirt-1(apo) and resveratrol-Sirt-1 docked complex. The docking of GBDG to the allosteric site along with favorable docking parameters, enhanced interactions between the catalytic and regulatory domains, increased complex stability (as shown by molecular dynamics simulation), and greater binding affinity to acetylated peptide substrate suggest that GBDG is a potent allosteric regulator of Sirt-1.
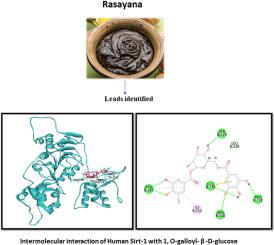
鉴定1- o -没食子酰-β- d -葡萄糖作为Sirtuin-1的有效激活剂:一项硅研究
Sirt-1是一种作用于组蛋白和多种非组蛋白的去乙酰化酶,在多种生理和病理过程中起着至关重要的作用。调节其活性的科学努力主要集中在小分子激活剂上。为了更好地鉴定激活Sirt-1的天然小分子化合物,本研究采用传统的知识驱动的基于选定药用植物的植物化学物质的计算机研究。利用植物化学文库对Sirt-1进行分子对接研究,发现1- o -没食子酰-β- d -葡萄糖(GBDG)与Sirt-1变构位点的对接评分、氢键相互作用等对接特征优于已知的Sirt-1天然小分子活化剂白藜芦醇。对对接物进行分子动力学模拟、轨迹分析(RMSD、RMSF、旋转半径和结合能),结果表明该配合物具有结构和热力学稳定性。调节域和催化域关键残基之间的质心距离测量表明,GBDG的对接导致催化域更靠近调节域的构象发生变化。与Sirt-1(apo)和白藜芦醇-Sirt-1对接的配合物相比,GBDG与Sirt-1的对接参数更好,表明GBDG与Sirt-1对接增加了其对乙酰化底物的亲和力。GBDG以良好的对接参数与变构位点对接,增强了催化和调控结构域之间的相互作用,增加了复合物的稳定性(如分子动力学模拟所示),并且与乙酰化肽底物的结合亲和力更强,这表明GBDG是Sirt-1的有效变构调节剂。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of molecular graphics & modelling
生物-计算机:跨学科应用
CiteScore
5.50
自引率
6.90%
发文量
216
审稿时长
35 days
期刊介绍:
The Journal of Molecular Graphics and Modelling is devoted to the publication of papers on the uses of computers in theoretical investigations of molecular structure, function, interaction, and design. The scope of the journal includes all aspects of molecular modeling and computational chemistry, including, for instance, the study of molecular shape and properties, molecular simulations, protein and polymer engineering, drug design, materials design, structure-activity and structure-property relationships, database mining, and compound library design.
As a primary research journal, JMGM seeks to bring new knowledge to the attention of our readers. As such, submissions to the journal need to not only report results, but must draw conclusions and explore implications of the work presented. Authors are strongly encouraged to bear this in mind when preparing manuscripts. Routine applications of standard modelling approaches, providing only very limited new scientific insight, will not meet our criteria for publication. Reproducibility of reported calculations is an important issue. Wherever possible, we urge authors to enhance their papers with Supplementary Data, for example, in QSAR studies machine-readable versions of molecular datasets or in the development of new force-field parameters versions of the topology and force field parameter files. Routine applications of existing methods that do not lead to genuinely new insight will not be considered.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


