Classification of glioma grade and Ki-67 level prediction in MRI data: A SHAP-driven interpretation
IF 4.9
2区 医学
Q1 ENGINEERING, BIOMEDICAL
Computerized Medical Imaging and Graphics
Pub Date : 2025-06-16
DOI:10.1016/j.compmedimag.2025.102578
引用次数: 0
Abstract
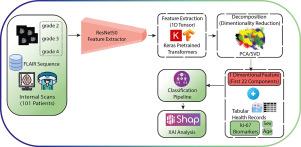
This study focuses on artificial intelligence-driven classification of glioma and Ki-67 leveling using T2w-FLAIR MRI, exploring the association of Ki-67 biomarkers with deep learning (DL) features through explainable artificial intelligence (XAI) and SHapley Additive exPlanations (SHAP). This IRB-approved study included 101 patients with glioma brain tumor acquired MR images with the T2W-FLAIR sequence. We extracted DL bottleneck features using ResNet50 from glioma MR images. Principal component analysis (PCA) was deployed for dimensionality reduction. XAI was used to identify potential features. The XGBosst classified the histologic grades of the glioma and the level of Ki-67. We integrated potential DL features with patient demographics (age and sex) and Ki-67 biomarkers, utilizing SHAP to determine the model’s essential features and interactions. Glioma grade classification and Ki-67 level predictions achieved overall accuracies of 0.94 and 0.91, respectively. It achieved precision scores of 0.92, 0.94, and 0.96 for glioma grades 2, 3, and 4, and 0.88, 0.94, and 0.97 for Ki-67 levels (low: , moderate: , and high: ). Corresponding F1-scores were 0.95, 0.88, and 0.96 for glioma grades and 0.92, 0.93, and 0.87 for Ki-67 levels. SHAP analysis further highlighted a strong association between bottleneck DL features and Ki-67 biomarkers, demonstrating their potential to differentiate glioma grades and Ki-67 levels while offering valuable insights into glioma aggressiveness. This study demonstrates the precise classification of glioma grades and the prediction of Ki-67 levels to underscore the potential of AI-driven MRI analysis to enhance clinical decision-making in glioma management.
MRI数据中胶质瘤分级和Ki-67水平预测的分类:shap驱动的解释
本研究的重点是使用T2w-FLAIR MRI进行人工智能驱动的胶质瘤分类和Ki-67水平,通过可解释人工智能(XAI)和SHapley加法解释(SHAP)探索Ki-67生物标志物与深度学习(DL)特征的关联。这项经irb批准的研究纳入了101例胶质瘤脑肿瘤患者,他们获得了T2W-FLAIR序列的MR图像。我们使用ResNet50从胶质瘤MR图像中提取DL瓶颈特征。采用主成分分析(PCA)进行降维。XAI用于识别潜在特征。XGBosst对胶质瘤的组织学分级和Ki-67水平进行分类。我们将潜在的深度学习特征与患者人口统计学(年龄和性别)和Ki-67生物标志物相结合,利用SHAP确定模型的基本特征和相互作用。胶质瘤分级和Ki-67水平预测的总体准确率分别为0.94和0.91。对于2级、3级和4级胶质瘤,其精确评分分别为0.92、0.94和0.96,对于Ki-67水平,其精确评分分别为0.88、0.94和0.97(低:5%≤Ki -67≤10%,中:10%≤Ki -67≤20,高:Ki-67≤20%)。胶质瘤分级对应的f1评分分别为0.95、0.88和0.96,Ki-67水平对应的f1评分分别为0.92、0.93和0.87。SHAP分析进一步强调了瓶颈DL特征与Ki-67生物标志物之间的强烈关联,证明了它们区分胶质瘤分级和Ki-67水平的潜力,同时为胶质瘤侵袭性提供了有价值的见解。本研究展示了胶质瘤分级的精确分类和Ki-67水平的预测,强调了人工智能驱动的MRI分析在增强胶质瘤管理的临床决策方面的潜力。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
CiteScore
10.70
自引率
3.50%
发文量
71
审稿时长
26 days
期刊介绍:
The purpose of the journal Computerized Medical Imaging and Graphics is to act as a source for the exchange of research results concerning algorithmic advances, development, and application of digital imaging in disease detection, diagnosis, intervention, prevention, precision medicine, and population health. Included in the journal will be articles on novel computerized imaging or visualization techniques, including artificial intelligence and machine learning, augmented reality for surgical planning and guidance, big biomedical data visualization, computer-aided diagnosis, computerized-robotic surgery, image-guided therapy, imaging scanning and reconstruction, mobile and tele-imaging, radiomics, and imaging integration and modeling with other information relevant to digital health. The types of biomedical imaging include: magnetic resonance, computed tomography, ultrasound, nuclear medicine, X-ray, microwave, optical and multi-photon microscopy, video and sensory imaging, and the convergence of biomedical images with other non-imaging datasets.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


