Improving diffusion-based protein backbone generation with global-geometry-aware latent encoding
IF 23.9
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
Abstract
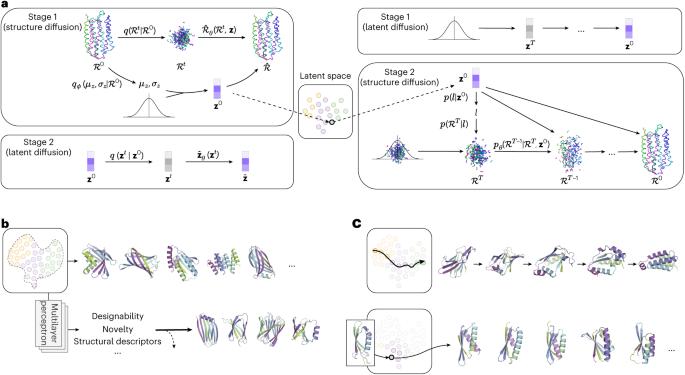
The global structural properties of a protein, such as shape, fold and topology, strongly affect its function. Although recent breakthroughs in diffusion-based generative models have greatly advanced de novo protein design, particularly in generating diverse and realistic structures, it remains challenging to design proteins of specific geometries without residue-level control over the topological details. A more practical, top-down approach is needed for prescribing the overall geometric arrangements of secondary structure elements in the generated protein structures. In response, we propose TopoDiff, an unsupervised framework that learns and exploits a global-geometry-aware latent representation, enabling both unconditional and controllable diffusion-based protein generation. Trained on the Protein Data Bank and CATH datasets, the structure encoder embeds protein global geometries into a 32-dimensional latent space, from which latent codes sampled by the latent sampler serve as informative conditions for the diffusion-based backbone decoder. In benchmarks against existing baselines, TopoDiff demonstrates comparable performance on established metrics including designability, diversity and novelty, as well as markedly improves coverage over the fold types of natural proteins in the CATH dataset. Moreover, latent conditioning enables versatile manipulations at the global-geometry level to control the generated protein structures, through which we derived a number of novel folds of mainly beta proteins with comprehensive experimental validation. A variational-autoencoder-based diffusion architecture that enables topological controls on the diffusion-based protein structure generation is proposed. As a result, novel folds of mainly beta proteins can be designed with experimental validation.
利用全局几何感知的隐编码改进基于扩散的蛋白质骨架生成
蛋白质的整体结构特性,如形状、折叠和拓扑结构,强烈影响其功能。尽管最近在基于扩散的生成模型方面的突破极大地推进了从头开始的蛋白质设计,特别是在生成多样化和逼真的结构方面,但在没有对拓扑细节进行残留级控制的情况下设计特定几何形状的蛋白质仍然具有挑战性。需要一种更实用的、自上而下的方法来规定生成的蛋白质结构中二级结构元素的总体几何排列。作为回应,我们提出了TopoDiff,这是一个无监督框架,可以学习和利用全局几何感知的潜在表示,从而实现无条件和可控的基于扩散的蛋白质生成。结构编码器在蛋白质数据库和CATH数据集上进行训练,将蛋白质全局几何形状嵌入到32维潜在空间中,潜在采样器从中采样的潜在代码作为基于扩散的骨干解码器的信息条件。在针对现有基线的基准测试中,TopoDiff在包括可设计性、多样性和新颖性在内的既定指标上表现出相当的性能,并显著提高了CATH数据集中天然蛋白质折叠类型的覆盖率。此外,潜在条件反射可以在全局几何水平上进行多种操作来控制生成的蛋白质结构,通过这种方法,我们获得了许多主要是β蛋白的新折叠,并得到了全面的实验验证。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Machine Intelligence
Multiple-
CiteScore
36.90
自引率
2.10%
发文量
127
期刊介绍:
Nature Machine Intelligence is a distinguished publication that presents original research and reviews on various topics in machine learning, robotics, and AI. Our focus extends beyond these fields, exploring their profound impact on other scientific disciplines, as well as societal and industrial aspects. We recognize limitless possibilities wherein machine intelligence can augment human capabilities and knowledge in domains like scientific exploration, healthcare, medical diagnostics, and the creation of safe and sustainable cities, transportation, and agriculture. Simultaneously, we acknowledge the emergence of ethical, social, and legal concerns due to the rapid pace of advancements.
To foster interdisciplinary discussions on these far-reaching implications, Nature Machine Intelligence serves as a platform for dialogue facilitated through Comments, News Features, News & Views articles, and Correspondence. Our goal is to encourage a comprehensive examination of these subjects.
Similar to all Nature-branded journals, Nature Machine Intelligence operates under the guidance of a team of skilled editors. We adhere to a fair and rigorous peer-review process, ensuring high standards of copy-editing and production, swift publication, and editorial independence.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


