Exploring novel insecticide candidates: Targeting Na+,K+ -ATPase in Drosophila melanogaster through computational and experimental approaches
IF 3
4区 生物学
Q2 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
Abstract
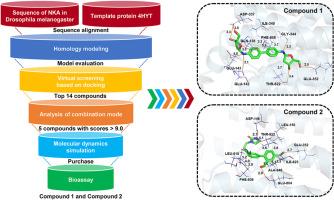
Pest infestations have been posing a serious and persistent threat to agricultural production. Traditional insecticides are currently confronted with a multitude of problems. The discovery of novel insecticide targets and the screening of active molecules can offer an entirely new direction for surmounting the limitations of traditional insecticides and lay a solid foundation for the development of highly effective insecticides with distinctive mechanisms of action. In this study, the Na+,K+-ATPase of Drosophila melanogaster has been selected as the target. Its three-dimensional structure has been constructed via homology modeling, and the evaluation has indicated that its quality is reliable. Subsequently, techniques including virtual screening, molecular docking, and molecular dynamics simulation have been employed to screen compounds and investigate their mechanisms of action. The analysis of binding modes has demonstrated that hydrogen bonding and hydrophobic interactions have played a crucial role in the binding of ligands. Molecular dynamics simulations and calculations of binding free energies have shown that Compound 1 and Compound 2 have exhibited similar or even stronger affinities in comparison to known inhibitors. Residue decomposition free energy reveals the types of key amino acid residues involved in the interaction between these compounds and NKA. Preliminary bioactivity assays have verified the bioactivities of these compounds. Ouabain, Compound 1, Compound 2, and Compound 4 have shown significant delayed toxic effects, with Compound 4 having a more pronounced delayed effect. Our study has provided certain valuable ideas and insights for the development of new insecticide molecules targeting the Na+,K+-ATPase of Drosophila melanogaster.
探索新的候选杀虫剂:通过计算和实验方法靶向黑腹果蝇的Na+,K+ - atp酶
病虫害一直对农业生产构成严重和持久的威胁。传统杀虫剂目前面临着许多问题。新的杀虫靶点的发现和活性分子的筛选可以为突破传统杀虫剂的局限性提供一个全新的方向,为开发具有独特作用机制的高效杀虫剂奠定坚实的基础。本研究选择黑腹果蝇的Na+,K+- atp酶作为靶点。通过同构建模构建了其三维结构,并对其质量进行了评价。随后,虚拟筛选、分子对接和分子动力学模拟等技术被用于筛选化合物并研究其作用机制。结合模式分析表明,氢键和疏水相互作用在配体的结合中起着至关重要的作用。分子动力学模拟和结合自由能计算表明,化合物1和化合物2与已知抑制剂具有相似甚至更强的亲和力。残基分解自由能揭示了这些化合物与NKA相互作用的关键氨基酸残基类型。初步的生物活性测定证实了这些化合物的生物活性。瓦巴因、化合物1、化合物2和化合物4均表现出明显的延迟毒性作用,其中化合物4的延迟效应更为明显。本研究为开发靶向果蝇Na+,K+- atp酶的新型杀虫分子提供了一定的思路和见解。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of molecular graphics & modelling
生物-计算机:跨学科应用
CiteScore
5.50
自引率
6.90%
发文量
216
审稿时长
35 days
期刊介绍:
The Journal of Molecular Graphics and Modelling is devoted to the publication of papers on the uses of computers in theoretical investigations of molecular structure, function, interaction, and design. The scope of the journal includes all aspects of molecular modeling and computational chemistry, including, for instance, the study of molecular shape and properties, molecular simulations, protein and polymer engineering, drug design, materials design, structure-activity and structure-property relationships, database mining, and compound library design.
As a primary research journal, JMGM seeks to bring new knowledge to the attention of our readers. As such, submissions to the journal need to not only report results, but must draw conclusions and explore implications of the work presented. Authors are strongly encouraged to bear this in mind when preparing manuscripts. Routine applications of standard modelling approaches, providing only very limited new scientific insight, will not meet our criteria for publication. Reproducibility of reported calculations is an important issue. Wherever possible, we urge authors to enhance their papers with Supplementary Data, for example, in QSAR studies machine-readable versions of molecular datasets or in the development of new force-field parameters versions of the topology and force field parameter files. Routine applications of existing methods that do not lead to genuinely new insight will not be considered.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


