Native structure of the monoclonal therapeutic CD20 antibody ocrelizumab
IF 2.3
4区 生物学
Q3 BIOCHEMISTRY & MOLECULAR BIOLOGY
Biochimica et biophysica acta. Proteins and proteomics
Pub Date : 2025-06-11
DOI:10.1016/j.bbapap.2025.141084
引用次数: 0
Abstract
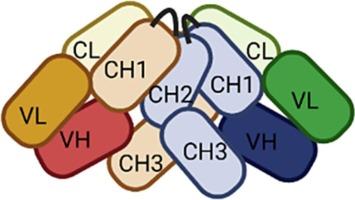
Immunoglobulin G (IgG) is fundamental to adaptive immunity and numerous monoclonal IgGs (monoclonal antibodies (MAbs)) have been developed as therapeutics for various diseases, including ocrelizumab (OMAb), a CD20 MAb used for treating multiple sclerosis, and infliximab (IMAb), a tumor necrosis factor MAb used for treating rheumatoid arthritis and other conditions. Understanding structure-function relationships are essential for understanding the mechanisms of action of IgG MAbs and previous results have shown that IgG has a “closed”, “m”-shaped conformation in native form, which may switch to an “open”, “Y”-shaped conformation upon antigen binding or physico-chemical stress. Supported by immunochemical and biophysical methods and by chemical crosslinking mass spectrometry (XL-MS) we show that both OMAb and IMAb conform to this paradigm. By XL-MS, we identified eighty-five high-confidence cross-links that support the native closed state of OMAb, refining our understanding of IgG architecture. Molecular modeling based on these data further corroborates a compact IgG structure, shielding the Fc domain. This structural insight may increase our understanding of immunoglobulin biology and enhance therapeutic MAb design by optimizing stability and efficacy.
单克隆治疗性CD20抗体ocrelizumab的天然结构
免疫球蛋白G (IgG)是适应性免疫的基础,许多单克隆IgG(单克隆抗体(MAb))已被开发用于各种疾病的治疗,包括ocrelizumab (OMAb),一种用于治疗多发性硬化症的CD20单抗,以及英夫利昔单抗(IMAb),一种用于治疗类风湿关节炎和其他疾病的肿瘤坏死因子单抗。了解结构-功能关系对于理解IgG单克隆抗体的作用机制至关重要,先前的研究结果表明,IgG在天然形态下具有“封闭”的“m”形构象,在抗原结合或物理化学胁迫下可能转变为“开放”的“Y”形构象。在免疫化学和生物物理方法以及化学交联质谱(XL-MS)的支持下,我们发现OMAb和IMAb都符合这一范式。通过xml - ms,我们确定了85个高置信度的交联,支持OMAb的原生封闭状态,完善了我们对IgG结构的理解。基于这些数据的分子模型进一步证实了一个紧凑的IgG结构,屏蔽了Fc结构域。这种结构的洞察力可能会增加我们对免疫球蛋白生物学的理解,并通过优化稳定性和有效性来增强治疗性单抗设计。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
CiteScore
8.00
自引率
0.00%
发文量
55
审稿时长
33 days
期刊介绍:
BBA Proteins and Proteomics covers protein structure conformation and dynamics; protein folding; protein-ligand interactions; enzyme mechanisms, models and kinetics; protein physical properties and spectroscopy; and proteomics and bioinformatics analyses of protein structure, protein function, or protein regulation.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


