Memory kernel minimization-based neural networks for discovering slow collective variables of biomolecular dynamics
IF 18.3
Q1 COMPUTER SCIENCE, INTERDISCIPLINARY APPLICATIONS
引用次数: 0
Abstract
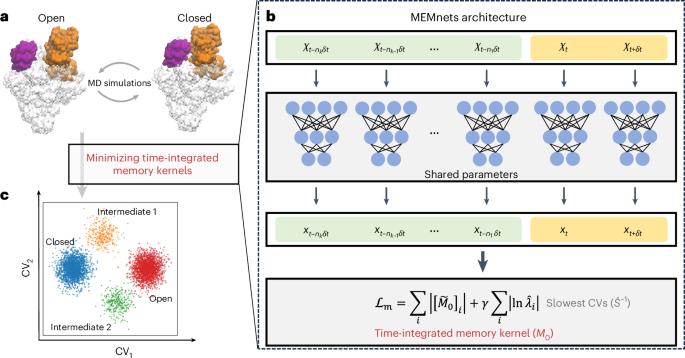
Identifying collective variables (CVs) that accurately capture the slowest timescales of protein conformational changes is crucial to comprehend numerous biological processes. Here we introduce memory kernel minimization-based neural networks (MEMnets), a deep learning framework that accurately identifies the slow CVs of biomolecular dynamics. Unlike popular CV-identification methods, which typically assume Markovian dynamics, MEMnets is built on the integrative generalized master equation theory, which incorporates non-Markovian dynamics by encoding them in a memory kernel for continuous CVs. The key innovation of MEMnets is the identification of optimal CVs by minimizing the upper bound for the time-integrated memory kernels through parallel encoder networks. We demonstrate that MEMnets effectively identifies slow CVs involved in the folding of the FIP35 WW domain, revealing two parallel folding pathways. In addition, we illustrate MEMnets’ robust numerical stability in identifying meaningful CVs in large biomolecular dynamic systems with limited sampling by applying it to the clamp opening of bacterial RNA polymerase, a much more complex conformational change. This study presents MEMnets, an approach that integrates statistical mechanics with deep learning to identify the slowest collective variables for biomolecular dynamics. MEMnets effectively captures memory effects, enabling advanced time-series analysis.
基于记忆核最小化的神经网络用于发现生物分子动力学的慢集体变量。
识别集体变量(cv),准确捕获蛋白质构象变化的最慢时间尺度,对于理解许多生物过程至关重要。在这里,我们介绍了基于记忆核最小化的神经网络(MEMnets),这是一种深度学习框架,可以准确识别生物分子动力学的慢cv。与传统的cv识别方法不同,MEMnets是建立在综合广义主方程理论的基础上的,该理论通过将非马尔可夫动力学编码到连续cv的记忆核中,从而将非马尔可夫动力学纳入其中。MEMnets的关键创新是通过并行编码器网络最小化时间积分记忆核的上界来识别最优cv。我们证明MEMnets有效地识别了参与FIP35 WW结构域折叠的慢速CVs,揭示了两个平行的折叠途径。此外,我们通过将MEMnets应用于细菌RNA聚合酶(一种更复杂的构象变化)的钳形打开,说明了MEMnets在有限采样的大型生物分子动力学系统中识别有意义的CVs的强大数值稳定性。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


