A multimodal conversational agent for DNA, RNA and protein tasks
IF 23.9
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
Abstract
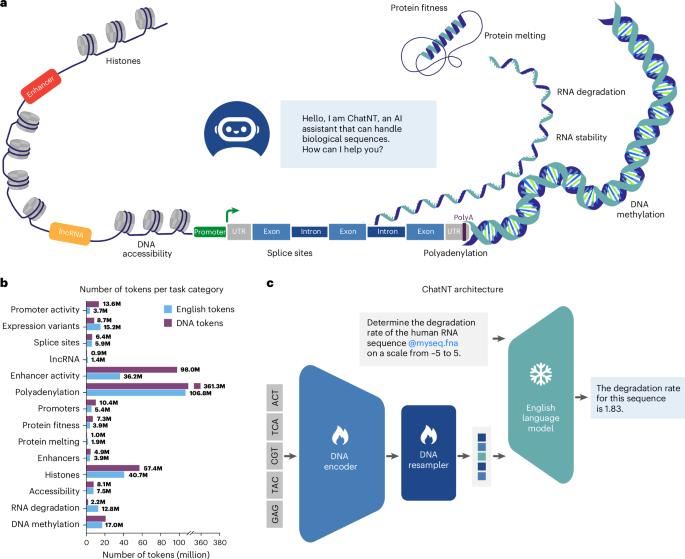
Language models are thriving, powering conversational agents that assist and empower humans to solve a number of tasks. Recently, these models were extended to support additional modalities including vision, audio and video, demonstrating impressive capabilities across multiple domains, including healthcare. Still, conversational agents remain limited in biology as they cannot yet fully comprehend biological sequences. Meanwhile, high-performance foundation models for biological sequences have been built through self-supervision over sequencing data, but these need to be fine-tuned for each specific application, preventing generalization between tasks. In addition, these models are not conversational, which limits their utility to users with coding capabilities. Here we propose to bridge the gap between biology foundation models and conversational agents by introducing ChatNT, a multimodal conversational agent with an advanced understanding of biological sequences. ChatNT achieves new state-of-the-art results on the Nucleotide Transformer benchmark while being able to solve all tasks at once, in English, and to generalize to unseen questions. In addition, we have curated a set of more biologically relevant instruction tasks from DNA, RNA and proteins, spanning multiple species, tissues and biological processes. ChatNT reaches performance on par with state-of-the-art specialized methods on those tasks. We also present a perplexity-based technique to help calibrate the confidence of our model predictions. By applying attribution methods through the English decoder and DNA encoder, we demonstrate that ChatNT’s answers are based on biologically coherent features such as detecting the promoter TATA motif or splice site dinucleotides. Our framework for genomics instruction tuning can be extended to more tasks and data modalities (for example, structure and imaging), making it a widely applicable tool for biology. ChatNT provides a potential direction for building generally capable agents that understand biology from first principles while being accessible to users with no coding background. De Almeida, Richard and colleagues leverage transfer learning to create ChatNT, a multimodal conversational agent for DNA, RNA and protein sequences that can be instructed in natural language.

DNA, RNA和蛋白质任务的多模式对话代理
语言模型正在蓬勃发展,为会话代理提供动力,帮助并赋予人类解决许多任务的能力。最近,这些模型进行了扩展,以支持包括视觉、音频和视频在内的其他模式,在包括医疗保健在内的多个领域展示了令人印象深刻的功能。然而,对话代理在生物学上仍然有限,因为它们还不能完全理解生物序列。同时,通过对测序数据的自我监督,已经建立了高性能的生物序列基础模型,但这些模型需要针对每个特定应用进行微调,从而阻碍了任务之间的泛化。此外,这些模型不是对话式的,这限制了它们对具有编码能力的用户的实用性。在这里,我们建议通过引入ChatNT来弥合生物学基础模型和会话代理之间的差距,ChatNT是一种对生物序列有高级理解的多模态会话代理。ChatNT在Nucleotide Transformer基准测试上取得了新的最先进的结果,同时能够一次性用英语解决所有任务,并归纳出未见过的问题。此外,我们已经策划了一套更多的生物学相关的指令任务,从DNA, RNA和蛋白质,跨越多个物种,组织和生物过程。ChatNT在这些任务上达到了与最先进的专门方法相当的性能。我们还提出了一种基于困惑的技术来帮助校准我们模型预测的置信度。通过英文解码器和DNA编码器应用归因方法,我们证明ChatNT的答案是基于生物学上一致的特征,如检测启动子TATA基序或剪接位点二核苷酸。我们的基因组学指令调整框架可以扩展到更多的任务和数据模式(例如,结构和成像),使其成为广泛适用于生物学的工具。ChatNT提供了一个潜在的方向,即构建具有一般能力的智能体,这些智能体可以从基本原理理解生物学,同时也可以被没有编码背景的用户访问。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Machine Intelligence
Multiple-
CiteScore
36.90
自引率
2.10%
发文量
127
期刊介绍:
Nature Machine Intelligence is a distinguished publication that presents original research and reviews on various topics in machine learning, robotics, and AI. Our focus extends beyond these fields, exploring their profound impact on other scientific disciplines, as well as societal and industrial aspects. We recognize limitless possibilities wherein machine intelligence can augment human capabilities and knowledge in domains like scientific exploration, healthcare, medical diagnostics, and the creation of safe and sustainable cities, transportation, and agriculture. Simultaneously, we acknowledge the emergence of ethical, social, and legal concerns due to the rapid pace of advancements.
To foster interdisciplinary discussions on these far-reaching implications, Nature Machine Intelligence serves as a platform for dialogue facilitated through Comments, News Features, News & Views articles, and Correspondence. Our goal is to encourage a comprehensive examination of these subjects.
Similar to all Nature-branded journals, Nature Machine Intelligence operates under the guidance of a team of skilled editors. We adhere to a fair and rigorous peer-review process, ensuring high standards of copy-editing and production, swift publication, and editorial independence.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


