A trajectory-informed model for detecting drug-drug-host interaction from real-world data
IF 4.5
2区 医学
Q2 COMPUTER SCIENCE, INTERDISCIPLINARY APPLICATIONS
引用次数: 0
Abstract
Objective
Adverse drug event (ADE) is a significant challenge to public health. Since data mining methods have been developed to identify signals of drug-drug interaction-induced (DDI-induced) or drug-host interaction-induced (DHI-induced) ADE from real-world data, we aim to develop a new method to detect adverse drug-drug interaction with a special awareness on patient characteristics.
Methods
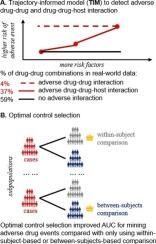
We developed a trajectory-informed model (TIM) to identify signals of adverse DDI with a special awareness on patient characteristics (i.e., drug-drug-host interaction [DDHI]). We also proposed a study design based on an optimal selection of within-subject and between-subjects controls for detecting ADEs from real-world data. We analyzed a large-scale US administrative claims data and conducted a simulation study.
Results
In administrative claims data analysis, we developed optimally matched case-control datasets for potential ADEs including acute kidney injury and gastrointestinal bleeding. We identified that an optimal selection of controls had a higher AUC compared to traditional designs for ADE detection (AUCs: 0.79–0.80 vs. 0.56–0.76). We observed that TIM detected more signals than reference methods (odds ratios: 1.13–3.18, P < 0.01), and found that 36 % of all signals generated by TIM were DDHI signals. In a simulation study, we demonstrated that TIM had an empirical false discovery rate (FDR) less than the desired value of 0.05, as well as > 1.4-fold higher probabilities of detection of DDHI signals than reference methods.
Conclusions
TIM had a high probability to identify signals of adverse DDI and DDHI in a high-throughput ADE mining while controlling false positive rate. A significant portion of drug-drug combinations were associated with an increased risk of ADEs only in specific patient subpopulations. Optimal selection of within-subject and between-subjects controls could improve the performance of ADE data mining.
从真实世界数据中检测药物-药物-宿主相互作用的轨迹知情模型
目的药物不良事件(ADE)是公共卫生面临的重大挑战。由于数据挖掘方法已经发展到从现实世界数据中识别药物-药物相互作用诱导(ddi诱导)或药物-宿主相互作用诱导(dhi诱导)ADE的信号,我们的目标是开发一种新的方法来检测药物-药物不良相互作用,同时对患者特征有特殊的认识。方法我们开发了一个轨迹信息模型(TIM)来识别不良的DDI信号,并对患者特征(即药物-药物-宿主相互作用[DDHI])有特殊的认识。我们还提出了一种基于受试者内和受试者间控制的最佳选择的研究设计,用于从现实世界数据中检测ade。我们分析了一项大规模的美国行政索赔数据并进行了模拟研究。结果在行政索赔数据分析中,我们开发了最优匹配的病例对照数据集,包括急性肾损伤和胃肠道出血。我们发现,与传统的ADE检测设计相比,最佳对照选择具有更高的AUC (AUC: 0.79-0.80 vs. 0.56-0.76)。我们观察到TIM比参考方法检测到更多的信号(优势比:1.13-3.18,P <;0.01),发现TIM产生的所有信号中36%为DDHI信号。在模拟研究中,我们证明了TIM的经验错误发现率(FDR)小于期望值0.05,并且>;检测DDHI信号的概率比参考方法高1.4倍。结论stim在高通量ADE挖掘中有较高的概率识别出不良DDI和DDHI信号,同时控制了假阳性率。很大一部分药物-药物联合用药仅在特定患者亚群中与ade风险增加相关。主题内和主题间控制的优化选择可以提高ADE数据挖掘的性能。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Biomedical Informatics
医学-计算机:跨学科应用
CiteScore
8.90
自引率
6.70%
发文量
243
审稿时长
32 days
期刊介绍:
The Journal of Biomedical Informatics reflects a commitment to high-quality original research papers, reviews, and commentaries in the area of biomedical informatics methodology. Although we publish articles motivated by applications in the biomedical sciences (for example, clinical medicine, health care, population health, and translational bioinformatics), the journal emphasizes reports of new methodologies and techniques that have general applicability and that form the basis for the evolving science of biomedical informatics. Articles on medical devices; evaluations of implemented systems (including clinical trials of information technologies); or papers that provide insight into a biological process, a specific disease, or treatment options would generally be more suitable for publication in other venues. Papers on applications of signal processing and image analysis are often more suitable for biomedical engineering journals or other informatics journals, although we do publish papers that emphasize the information management and knowledge representation/modeling issues that arise in the storage and use of biological signals and images. System descriptions are welcome if they illustrate and substantiate the underlying methodology that is the principal focus of the report and an effort is made to address the generalizability and/or range of application of that methodology. Note also that, given the international nature of JBI, papers that deal with specific languages other than English, or with country-specific health systems or approaches, are acceptable for JBI only if they offer generalizable lessons that are relevant to the broad JBI readership, regardless of their country, language, culture, or health system.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


