Interpretable niche-based cell‒cell communication inference using multi-view graph neural networks
IF 18.3
Q1 COMPUTER SCIENCE, INTERDISCIPLINARY APPLICATIONS
引用次数: 0
Abstract
Cell‒cell communication (CCC) is a fundamental biological process for the harmonious functioning of biological systems. Increasing evidence indicates that cells of the same type or cluster may exhibit different interaction patterns under varying niches, yet most prevailing methods perform CCC inference at the cell type or cluster level while disregarding niche heterogeneity. Here we introduce the Spatial Transcriptomics-based cell‒cell Communication And Subtype Exploration (STCase) tool, which can describe CCC events at the single-cell/spot level based on spatial transcriptomics (ST). STCase includes an interpretable multi-view graph neural network via CCC-aware attention to identify niches for each cell type and uncover niche-specific CCC events. We show that STCase outperforms state-of-the-art approaches and accurately captures reported immune-related CCC events in human bronchial glands. We also identify three distinct niches of oral squamous cell carcinoma that may be obscured by agglomerative methods, and discover niche-specific CCC events that could influence tumor prognosis. An interpretable tool called STCase is introduced to leverage a multi-view graph neural network based on cell‒cell communication (CCC)-aware attention to uncover functional niche-specific CCC events based on spatial transcriptomics data.
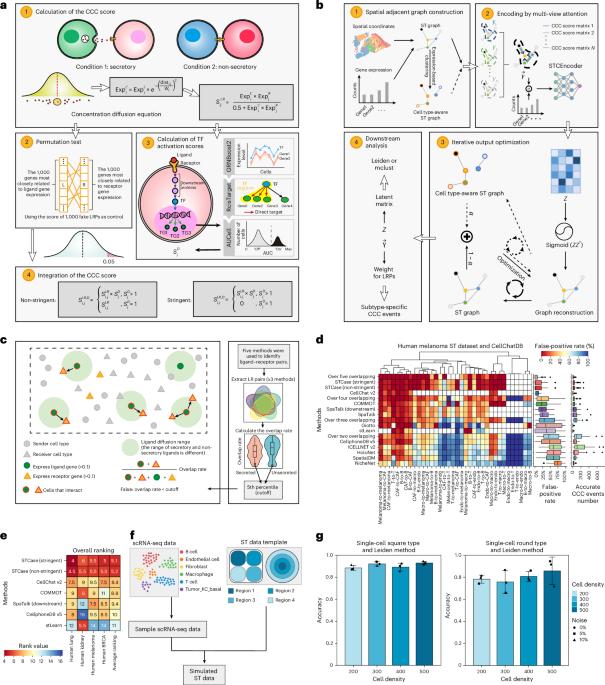
利用多视图图神经网络可解释的基于小生境的细胞-细胞通信推理。
细胞间通讯是生物系统和谐运行的基本生物学过程。越来越多的证据表明,相同类型或集群的细胞在不同的生态位下可能表现出不同的相互作用模式,但大多数流行的方法在细胞类型或集群水平上进行CCC推理,而忽略了生态位异质性。在这里,我们介绍了基于空间转录组学的细胞-细胞通信和亚型探索(STCase)工具,该工具可以基于空间转录组学(ST)描述单细胞/斑点水平的CCC事件。STCase包括一个可解释的多视图神经网络,通过CCC感知注意力来识别每种细胞类型的利基,并揭示利基特定的CCC事件。我们表明,STCase优于最先进的方法,并准确捕获报告的人类支气管腺中免疫相关的CCC事件。我们还确定了口腔鳞状细胞癌的三个不同的利基,这些利基可能被聚集方法所掩盖,并发现了可能影响肿瘤预后的利基特异性CCC事件。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


