Impacts of sample weighting on transferability of risk prediction models across EHR-Linked biobanks with different recruitment strategies
IF 4.5
2区 医学
Q2 COMPUTER SCIENCE, INTERDISCIPLINARY APPLICATIONS
引用次数: 0
Abstract
Objective
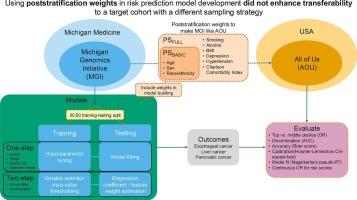
To evaluate whether using poststratification weights when training risk prediction models enhances transferability when the external test cohort has a different sampling strategy, a commonly encountered scenario when analyzing electronic health record (EHR)-linked biobanks.
Methods
PS weights were calculated to align a health system-based biobank, the Michigan Genomics Initiative (MGI; n = 76,757), with a nationally recruited biobank, All of Us (AOU; n = 226,764), which oversamples underrepresented groups. Basic PS weights (PSBASIC) captured age, sex, and race/ethnicity; full PS weights (PSFULL) additionally included smoking, alcohol consumption, BMI, depression, hypertension, and the Charlson Comorbidity Index. Models for esophageal, liver, and pancreatic cancers were developed using EHR data from MGI at 0, 1, 2, and 5 years prior to diagnosis. Phenotype risk scores (PheRS) were constructed using six methods (e.g., regularized regression, random forest) and evaluated alongside covariates, risk factors, and symptoms. Evaluation metrics included the odds ratio (OR) for the top decile vs. the middle 40th-60th percentiles of the risk score distribution and the area under the receiver operating curve (AUC) evaluated in the AOU test cohort when models are trained with and without weighting.
Results
Elastic net and random forest methods generally performed well in risk stratification, but no single PheRS construction method consistently outperformed others. Applying PS weights did not consistently improve risk stratification performance. For example, in liver cancer risk stratification at t = 1, unweighted random forest PheRS yielded an OR of 13.73 (95 % CI: 8.97, 21.01), compared to 14.55 (95 % CI: 9.45, 22.42) with PSBASIC and 13.62 (95 % CI: 8.90, 20.85) with PSFULL.
Conclusion
PS weights do not significantly enhance risk model transferability between biobanks. EHR-based PheRS are crucial for risk stratification and should be integrated with other multimodal data for improved risk prediction. Identifying high-risk populations for diseases like liver cancer early through health history mining shows promise.
样本权重对不同招募策略下ehr关联生物库风险预测模型可转移性的影响
目的评估当外部测试队列采用不同的抽样策略时,在训练风险预测模型时使用分层后权重是否能增强可转移性,这是分析电子健康记录(EHR)相关生物库时经常遇到的情况。方法计算sps的权重,以对齐基于卫生系统的生物库,密歇根基因组计划(MGI;n = 76,757),与全国招募的生物库,我们所有人(AOU;N = 226,764),这是对代表性不足群体的过度抽样。基本PS权重(PSBASIC)捕获年龄、性别和种族/民族;全PS体重(PSFULL)还包括吸烟、饮酒、BMI、抑郁、高血压和Charlson合并症指数。食管癌、肝癌和胰腺癌的模型是利用MGI在诊断前0、1、2和5年的电子病历数据开发的。表型风险评分(PheRS)采用六种方法(如正则化回归、随机森林)构建,并与协变量、危险因素和症状一起评估。评估指标包括风险评分分布的前十分位数与中间40 -60百分位数的比值比(OR),以及在AOU测试队列中对模型进行加权和不加权训练时评估的受试者操作曲线下面积(AUC)。结果弹性网法和随机森林法在风险分层中普遍表现较好,但没有单一的PheRS构建方法具有一致性。应用PS权重并不能持续改善风险分层的表现。例如,在t = 1的肝癌风险分层中,未加权随机森林PheRS的OR为13.73 (95% CI: 8.97, 21.01),而PSBASIC的OR为14.55 (95% CI: 9.45, 22.42), PSFULL的OR为13.62 (95% CI: 8.90, 20.85)。结论ps权重不能显著提高风险模型在生物库之间的可转移性。基于ehr的PheRS对风险分层至关重要,应与其他多模式数据相结合,以改进风险预测。通过健康史挖掘,及早发现肝癌等疾病的高危人群,这是有希望的。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Biomedical Informatics
医学-计算机:跨学科应用
CiteScore
8.90
自引率
6.70%
发文量
243
审稿时长
32 days
期刊介绍:
The Journal of Biomedical Informatics reflects a commitment to high-quality original research papers, reviews, and commentaries in the area of biomedical informatics methodology. Although we publish articles motivated by applications in the biomedical sciences (for example, clinical medicine, health care, population health, and translational bioinformatics), the journal emphasizes reports of new methodologies and techniques that have general applicability and that form the basis for the evolving science of biomedical informatics. Articles on medical devices; evaluations of implemented systems (including clinical trials of information technologies); or papers that provide insight into a biological process, a specific disease, or treatment options would generally be more suitable for publication in other venues. Papers on applications of signal processing and image analysis are often more suitable for biomedical engineering journals or other informatics journals, although we do publish papers that emphasize the information management and knowledge representation/modeling issues that arise in the storage and use of biological signals and images. System descriptions are welcome if they illustrate and substantiate the underlying methodology that is the principal focus of the report and an effort is made to address the generalizability and/or range of application of that methodology. Note also that, given the international nature of JBI, papers that deal with specific languages other than English, or with country-specific health systems or approaches, are acceptable for JBI only if they offer generalizable lessons that are relevant to the broad JBI readership, regardless of their country, language, culture, or health system.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


