Physics-based modeling in the new era of enzyme engineering
IF 18.3
Q1 COMPUTER SCIENCE, INTERDISCIPLINARY APPLICATIONS
引用次数: 0
Abstract
Enzyme engineering is entering a new era characterized by the integration of computational strategies. While bioinformatics and artificial intelligence methods have been extensively applied to accelerate the screening of function-enhancing mutants, physics-based modeling methods, such as molecular mechanics and quantum mechanics, are essential complements in many objectives. In this Perspective, we highlight how physics-based modeling will help the field of computational enzyme engineering reach its full potential by exploring current developments, unmet challenges and emerging opportunities for tool development. This Perspective highlights the vital role of physics-based modeling in computational enzyme engineering, exploring key advances, challenges and future steps. By integrating machine learning, these approaches can enhance each other, unlocking the full potential of enzyme design and discovery.
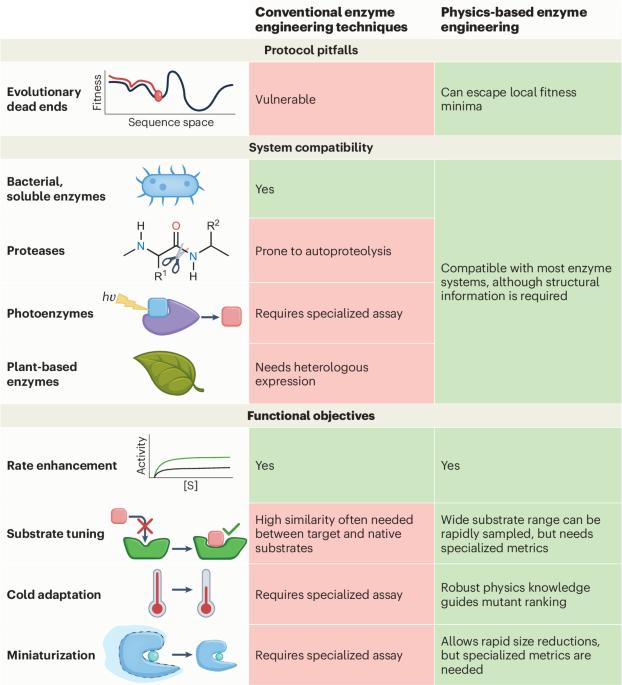
酶工程新时代的物理建模。
酶工程正在进入一个以计算策略集成为特征的新时代。虽然生物信息学和人工智能方法已被广泛应用于加速功能增强突变体的筛选,但基于物理的建模方法,如分子力学和量子力学,在许多目标中是必不可少的补充。在这个视角中,我们强调了基于物理的建模将如何通过探索当前的发展、未遇到的挑战和工具开发的新机遇来帮助计算酶工程领域发挥其全部潜力。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


