A lightweight graph neural network to predict long-term mortality in coronary artery disease patients: an interpretable causality-aware approach
IF 4.5
2区 医学
Q2 COMPUTER SCIENCE, INTERDISCIPLINARY APPLICATIONS
引用次数: 0
Abstract
Background
Coronary artery disease (CAD) causes substantial death toll in the United States and worldwide. While traditional methods for CAD mortality prediction are based on established risk factors, they have significant limitations in accuracy, adaptability to diverse populations, performance for individual risk prediction compared to group data, and incorporation of socioeconomic and lifestyle variations. Machine learning (ML) models have demonstrated superior performance in CAD prediction; however, they often struggle with capturing complex data interactions that can impact mortality.
Methods
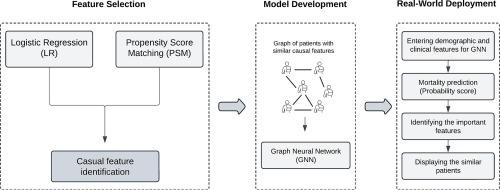
We proposed lightweight, interpretable graph neural network (GNN) models, utilizing data from a large trial of hypertensive patients with CAD to predict mortality using a concise set of critical features. While this smaller set of features can improve efficiency and implementation in clinical settings, the model’s “lightweight” nature facilitates fast real-time applications. We utilized a hybrid approach, which first uses logistic regression (LR) to identify statistically significant features, followed by propensity score matching (PSM) to identify potentially causal features. These causal features, alongside demographic variables, were employed to create a graph of patients, drawing edges between patients with similar causal features. Accordingly, lightweight 5-layer graph convolutional network (GCN) and graph attention network (GAT) were designed for mortality prediction, followed by an interpretable method (i.e., GNNExplainer) to report the feature importance.
Results
The proposed GCN achieved a recall of 93.02% and a negative predictive value (NPV) of 89.42%, higher than all other classifiers. Accordingly, a web-based decision support system (DSS), called CAD-SS, was developed, capable of predicting mortality and identifying risk factors and similar patients, guiding clinicians in reliable and informed decision-making.
Conclusions
Our proposed CAD-SS, which utilizes an interpretable and causality-aware lightweight GCN model, demonstrated reasonably high performance in predicting mortality due to CAD. This unique system can help identify the most vulnerable patients.
预测冠心病患者长期死亡率的轻量级图神经网络:一种可解释的因果关系感知方法。
背景:冠状动脉疾病(CAD)在美国和世界范围内造成大量死亡。虽然传统的CAD死亡率预测方法是基于既定的危险因素,但它们在准确性、对不同人群的适应性、与群体数据相比的个体风险预测性能以及社会经济和生活方式变化方面存在显着局限性。机器学习(ML)模型在CAD预测中表现出优异的性能;然而,他们常常难以捕捉到可能影响死亡率的复杂数据交互。方法:我们提出了轻量级、可解释的图神经网络(GNN)模型,利用一项大型高血压CAD患者试验的数据,使用一组简明的关键特征来预测死亡率。虽然这种较小的功能集可以提高临床环境中的效率和实施,但该模型的“轻量级”性质有助于快速实时应用。我们采用了一种混合方法,首先使用逻辑回归(LR)来识别统计上显著的特征,然后使用倾向评分匹配(PSM)来识别潜在的因果特征。这些因果特征与人口统计变量一起被用来创建一个患者图,在具有相似因果特征的患者之间画出边缘。因此,设计轻量级5层图卷积网络(GCN)和图注意网络(GAT)进行死亡率预测,然后采用可解释方法(即gnexplinterpreter)报告特征重要性。结果:GCN的召回率为93.02 %,负预测值(NPV)为89.42 %,高于所有其他分类器。因此,开发了基于网络的决策支持系统(DSS),称为CAD-SS,能够预测死亡率,识别风险因素和类似患者,指导临床医生做出可靠和知情的决策。结论:我们提出的CAD- ss,利用可解释和因果关系感知的轻量级GCN模型,在预测CAD死亡率方面表现出相当高的性能。这个独特的系统可以帮助识别最脆弱的病人。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Biomedical Informatics
医学-计算机:跨学科应用
CiteScore
8.90
自引率
6.70%
发文量
243
审稿时长
32 days
期刊介绍:
The Journal of Biomedical Informatics reflects a commitment to high-quality original research papers, reviews, and commentaries in the area of biomedical informatics methodology. Although we publish articles motivated by applications in the biomedical sciences (for example, clinical medicine, health care, population health, and translational bioinformatics), the journal emphasizes reports of new methodologies and techniques that have general applicability and that form the basis for the evolving science of biomedical informatics. Articles on medical devices; evaluations of implemented systems (including clinical trials of information technologies); or papers that provide insight into a biological process, a specific disease, or treatment options would generally be more suitable for publication in other venues. Papers on applications of signal processing and image analysis are often more suitable for biomedical engineering journals or other informatics journals, although we do publish papers that emphasize the information management and knowledge representation/modeling issues that arise in the storage and use of biological signals and images. System descriptions are welcome if they illustrate and substantiate the underlying methodology that is the principal focus of the report and an effort is made to address the generalizability and/or range of application of that methodology. Note also that, given the international nature of JBI, papers that deal with specific languages other than English, or with country-specific health systems or approaches, are acceptable for JBI only if they offer generalizable lessons that are relevant to the broad JBI readership, regardless of their country, language, culture, or health system.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


