Knowledge-enhanced Parameter-efficient Transfer Learning with METER for medical vision-language tasks
IF 4.5
2区 医学
Q2 COMPUTER SCIENCE, INTERDISCIPLINARY APPLICATIONS
引用次数: 0
Abstract
Objective:
The full fine-tuning paradigm becomes impractical when applying pre-trained models to downstream tasks due to significant computational and storage costs. Parameter-efficient fine-tuning (PEFT) methods can alleviate the issue. However, solely applying PEFT methods leads to sub-optimal performance owing to the domain gap between pre-trained models and medical downstream tasks.
Methods:
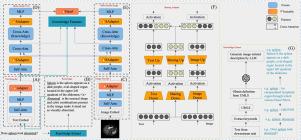
This study proposes Knowledge-enhanced Parameter-efficient Transfer Learning with METER (KPL-METER) for medical vision-language (VL) downstream tasks. KPL-METER combines PEFT methods, including an innovative PEFT module for multi-modal branches and newly introduced external domain-specific knowledge to enhance model performance. First, a lightweight, plug-and-play module named Sharing Adapter (SAdapter) is developed and inserted into the multi-modal encoders. This allows the two modalities to maintain uni-modal features while encouraging cross-modal consistency. Second, a novel knowledge extraction method and a parameter-free knowledge modeling strategy are developed to incorporate domain-specific knowledge from the Unified Medical Language System (UMLS) into multi-modal features. To further enhance the modeling of uni-modal features, Adapter is added to the image and text encoders.
Results:
The effectiveness of the proposed model is evaluated on two medical VL tasks using three VL datasets. The results indicate that the KPL-METER model outperforms other PEFT methods in terms of performance while utilizing fewer parameters. Furthermore, KPL-METER-MED, which incorporates medical-tailored encoders, is developed. Compared to previous models in the medical domain, KPL-METER-MED tunes fewer parameters while generally achieving higher performance.
Conclusion:
The proposed KPL-METER architecture effectively adapts general VL models for medical VL tasks, and the designed knowledge extraction and fusion method notably enhance performance by integrating medical domain-specific knowledge. Code is available at https://github.com/Adam-lxd/KPL-METER.
基于METER的医学视觉语言任务的知识增强参数高效迁移学习
目的:由于显著的计算和存储成本,当将预训练模型应用于下游任务时,完全微调范式变得不切实际。参数有效微调(PEFT)方法可以缓解这一问题。然而,由于预训练模型和医疗下游任务之间的域差距,单独应用PEFT方法会导致次优性能。方法:本研究提出基于METER的知识增强参数高效迁移学习方法(KPL-METER)用于医学视觉语言(VL)下游任务。KPL-METER结合了PEFT方法,包括用于多模态分支的创新PEFT模块和新引入的外部领域特定知识,以增强模型性能。首先,开发了一个名为共享适配器(SAdapter)的轻量级即插即用模块,并将其插入到多模态编码器中。这允许两种模态保持单模态特性,同时鼓励跨模态一致性。其次,提出了一种新的知识提取方法和无参数知识建模策略,将统一医学语言系统(Unified Medical Language System, UMLS)中的特定领域知识整合到多模态特征中;为了进一步增强单模态特征的建模,适配器被添加到图像和文本编码器中。结果:使用三个VL数据集评估了该模型在两个医疗VL任务上的有效性。结果表明,KPL-METER模型在使用较少参数的情况下,在性能方面优于其他PEFT方法。此外,还开发了KPL-METER-MED,其中包含医疗定制编码器。与医学领域以前的模型相比,KPL-METER-MED调整的参数更少,但通常可以实现更高的性能。结论:本文提出的KPL-METER体系结构能有效地适应医学VL任务的通用VL模型,所设计的知识提取与融合方法通过整合医学领域特定知识,显著提高了性能。代码可从https://github.com/Adam-lxd/KPL-METER获得。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Biomedical Informatics
医学-计算机:跨学科应用
CiteScore
8.90
自引率
6.70%
发文量
243
审稿时长
32 days
期刊介绍:
The Journal of Biomedical Informatics reflects a commitment to high-quality original research papers, reviews, and commentaries in the area of biomedical informatics methodology. Although we publish articles motivated by applications in the biomedical sciences (for example, clinical medicine, health care, population health, and translational bioinformatics), the journal emphasizes reports of new methodologies and techniques that have general applicability and that form the basis for the evolving science of biomedical informatics. Articles on medical devices; evaluations of implemented systems (including clinical trials of information technologies); or papers that provide insight into a biological process, a specific disease, or treatment options would generally be more suitable for publication in other venues. Papers on applications of signal processing and image analysis are often more suitable for biomedical engineering journals or other informatics journals, although we do publish papers that emphasize the information management and knowledge representation/modeling issues that arise in the storage and use of biological signals and images. System descriptions are welcome if they illustrate and substantiate the underlying methodology that is the principal focus of the report and an effort is made to address the generalizability and/or range of application of that methodology. Note also that, given the international nature of JBI, papers that deal with specific languages other than English, or with country-specific health systems or approaches, are acceptable for JBI only if they offer generalizable lessons that are relevant to the broad JBI readership, regardless of their country, language, culture, or health system.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


