Machine learning prediction of enzyme optimum pH
IF 23.9
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
Abstract
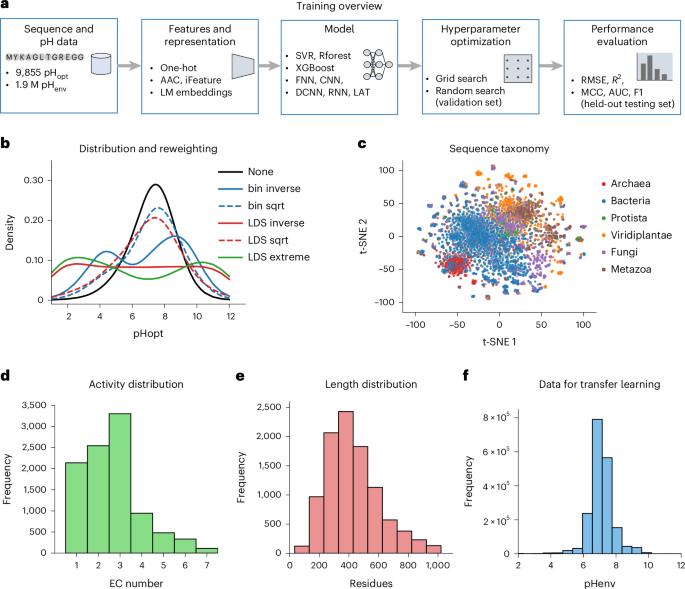
The relationship between pH and enzyme catalytic activity, especially the optimal pH (pHopt) at which enzymes function, is critical for biotechnological applications. Hence, computational methods to predict pHopt will enhance enzyme discovery and design by facilitating accurate identification of enzymes that function optimally at specific pH levels, and by elucidating sequence–function relationships. Here we proposed and evaluated various machine learning methods for predicting pHopt, conducting extensive hyperparameter optimization and training over 11,000 model instances. Our results demonstrate that models utilizing language model embeddings markedly outperform other methods in predicting pHopt. We present EpHod, the best-performing model, to predict pHopt, making it publicly available to researchers. From sequence data, EpHod directly learns structural and biophysical features that relate to pHopt, including proximity of residues to the catalytic centre and the accessibility of solvent molecules. Overall, EpHod presents a promising advancement in pHopt prediction and will potentially speed up the development of enzyme technologies. Accurately predicting the optimal pH level for enzyme activity is challenging due to the complex relationship between enzyme structure and function. Gado and colleagues show that a language model can effectively learn the structural and biophysical features to predict the optimal pH for enzyme activity.

酶最适pH的机器学习预测
pH值与酶的催化活性之间的关系,特别是酶发挥作用的最佳pH值(pHopt),对生物技术应用至关重要。因此,预测pHopt的计算方法将通过促进准确识别在特定pH水平下功能最佳的酶,并通过阐明序列-功能关系,来增强酶的发现和设计。在这里,我们提出并评估了预测pHopt的各种机器学习方法,进行了广泛的超参数优化和训练超过11,000个模型实例。我们的研究结果表明,使用语言模型嵌入的模型在预测pHopt方面明显优于其他方法。我们提出了预测pHopt的最佳模型EpHod,并将其公开提供给研究人员。从序列数据中,EpHod直接了解到与pHopt相关的结构和生物物理特征,包括残基与催化中心的接近程度和溶剂分子的可及性。总的来说,EpHod在pHopt预测方面有很大的进步,并有可能加速酶技术的发展。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Machine Intelligence
Multiple-
CiteScore
36.90
自引率
2.10%
发文量
127
期刊介绍:
Nature Machine Intelligence is a distinguished publication that presents original research and reviews on various topics in machine learning, robotics, and AI. Our focus extends beyond these fields, exploring their profound impact on other scientific disciplines, as well as societal and industrial aspects. We recognize limitless possibilities wherein machine intelligence can augment human capabilities and knowledge in domains like scientific exploration, healthcare, medical diagnostics, and the creation of safe and sustainable cities, transportation, and agriculture. Simultaneously, we acknowledge the emergence of ethical, social, and legal concerns due to the rapid pace of advancements.
To foster interdisciplinary discussions on these far-reaching implications, Nature Machine Intelligence serves as a platform for dialogue facilitated through Comments, News Features, News & Views articles, and Correspondence. Our goal is to encourage a comprehensive examination of these subjects.
Similar to all Nature-branded journals, Nature Machine Intelligence operates under the guidance of a team of skilled editors. We adhere to a fair and rigorous peer-review process, ensuring high standards of copy-editing and production, swift publication, and editorial independence.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


