Exportin-1 functions as an adaptor for transcription factor-mediated docking of chromatin at the nuclear pore complex
IF 14.5
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
Nuclear pore proteins (nucleoporins [Nups]) physically interact with hundreds of chromosomal sites, impacting transcription. In yeast, transcription factors mediate interactions between Nups and enhancers and promoters. To define the molecular basis of this mechanism, we exploited a separation-of-function mutation in the Gcn4 transcription factor that blocks its interaction with the nuclear pore complex (NPC). This mutation reduces the interaction of Gcn4 with the highly conserved nuclear export factor Crm1/Xpo1. Crm1 and Nups co-occupy enhancers, and Crm1 inhibition blocks interaction of the nuclear pore protein Nup2 with the genome. In vivo, Crm1 interacts stably with the NPC and in vitro, Crm1 binds directly to both Gcn4 and Nup2. Importantly, the interaction between Crm1 and Gcn4 requires neither Ran-guanosine triphosphate (GTP) nor the nuclear export sequence binding site. Finally, Crm1 and Ran-GTP stimulate DNA binding by Gcn4, suggesting that allosteric coupling between Crm1-Ran-GTP binding and DNA binding facilitates the docking of transcription-factor-bound enhancers at the NPC.
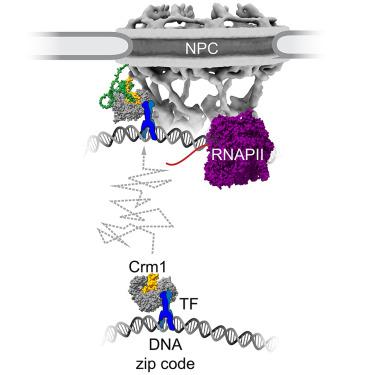
export -1作为转录因子介导的核孔复合物染色质对接的接头
核孔蛋白(核孔蛋白[Nups])物理上与数百个染色体位点相互作用,影响转录。在酵母中,转录因子介导nup与增强子和启动子之间的相互作用。为了确定这一机制的分子基础,我们利用了Gcn4转录因子中的一个功能分离突变,该突变阻断了其与核孔复合物(NPC)的相互作用。这种突变减少了Gcn4与高度保守的核输出因子Crm1/Xpo1的相互作用。Crm1和Nups共占据增强子,Crm1抑制可阻断核孔蛋白Nup2与基因组的相互作用。在体内,Crm1与NPC稳定地相互作用,在体外,Crm1直接与Gcn4和Nup2结合。重要的是,Crm1和Gcn4之间的相互作用既不需要三磷酸甘鸟苷(GTP),也不需要核输出序列结合位点。最后,Crm1和Ran-GTP通过Gcn4刺激DNA结合,表明Crm1-Ran-GTP结合与DNA结合之间的变构耦合促进了转录因子结合增强子在NPC的对接。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Molecular Cell
生物-生化与分子生物学
CiteScore
26.00
自引率
3.80%
发文量
389
审稿时长
1 months
期刊介绍:
Molecular Cell is a companion to Cell, the leading journal of biology and the highest-impact journal in the world. Launched in December 1997 and published monthly. Molecular Cell is dedicated to publishing cutting-edge research in molecular biology, focusing on fundamental cellular processes. The journal encompasses a wide range of topics, including DNA replication, recombination, and repair; Chromatin biology and genome organization; Transcription; RNA processing and decay; Non-coding RNA function; Translation; Protein folding, modification, and quality control; Signal transduction pathways; Cell cycle and checkpoints; Cell death; Autophagy; Metabolism.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


