Integrating epidemiology and genomics data to estimate the prevalence of acquired cysteine drug targets in the U.S. cancer patient population
IF 2.9
3区 医学
Q2 GENETICS & HEREDITY
引用次数: 0
Abstract
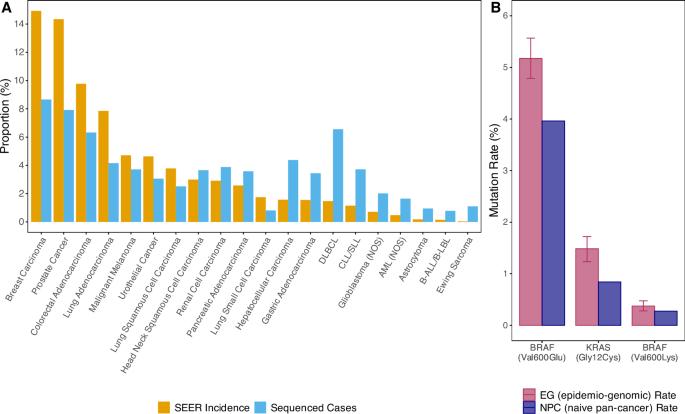
Reliable estimates for the number of cancer patients with a specific mutation can help quantify the size of the population that could potentially benefit from a targeted therapy. We adapt our previously developed approach for estimating gene-level mutation abundances to estimate mutation-specific (e.g., KRAS G12C) abundances by combining United States cancer epidemiology and genomic data. We demonstrate the approach by obtaining population-level estimates for all acquired somatic missense mutations that create a de novo cysteine residue. We find that approximately 14% of non-epidemiological informed estimates are more than twice the epidemiological informed estimate. Non-epidemiologically informed pan-cancer estimation of mutation rates may not be representative of the number of cancer patients with a specific mutation. Our study suggests that epidemiological and genomic information should be combined when estimating the population level abundance of specific pathogenic mutations.
整合流行病学和基因组学数据来估计获得性半胱氨酸药物靶点在美国癌症患者群体中的流行程度。
对具有特定突变的癌症患者数量的可靠估计可以帮助量化可能从靶向治疗中受益的人群的规模。通过结合美国癌症流行病学和基因组数据,我们调整了之前开发的估计基因水平突变丰度的方法,以估计突变特异性(例如,KRAS G12C)丰度。我们通过获得所有获得的产生新生半胱氨酸残基的体细胞错义突变的群体水平估计来证明该方法。我们发现,约14%的非流行病学知情估计是流行病学知情估计的两倍以上。没有流行病学信息的泛癌症突变率估计可能不能代表具有特定突变的癌症患者的数量。我们的研究表明,在估计特定致病突变的种群水平丰度时,应结合流行病学和基因组信息。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Pharmacogenomics Journal
医学-药学
CiteScore
7.20
自引率
0.00%
发文量
35
审稿时长
6-12 weeks
期刊介绍:
The Pharmacogenomics Journal is a print and electronic journal, which is dedicated to the rapid publication of original research on pharmacogenomics and its clinical applications.
Key areas of coverage include:
Personalized medicine
Effects of genetic variability on drug toxicity and efficacy
Identification and functional characterization of polymorphisms relevant to drug action
Pharmacodynamic and pharmacokinetic variations and drug efficacy
Integration of new developments in the genome project and proteomics into clinical medicine, pharmacology, and therapeutics
Clinical applications of genomic science
Identification of novel genomic targets for drug development
Potential benefits of pharmacogenomics.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


