Large language models for scientific discovery in molecular property prediction
IF 18.8
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
Abstract
Large language models (LLMs) are a form of artificial intelligence system encapsulating vast knowledge in the form of natural language. These systems are adept at numerous complex tasks including creative writing, storytelling, translation, question-answering, summarization and computer code generation. Although LLMs have seen initial applications in natural sciences, their potential for driving scientific discovery remains largely unexplored. In this work, we introduce LLM4SD, a framework designed to harness LLMs for driving scientific discovery in molecular property prediction by synthesizing knowledge from literature and inferring knowledge from scientific data. LLMs synthesize knowledge by extracting established information from scientific literature, such as molecular weight being key to predicting solubility. For inference, LLMs identify patterns in molecular data, particularly in Simplified Molecular Input Line Entry System-encoded structures, such as halogen-containing molecules being more likely to cross the blood–brain barrier. This information is presented as interpretable knowledge, enabling the transformation of molecules into feature vectors. By using these features with interpretable models such as random forest, LLM4SD can outperform the current state of the art across a range of benchmark tasks for predicting molecular properties. We foresee it providing interpretable and potentially new insights, aiding scientific discovery in molecular property prediction. Zheng et al. developed LLM4SD, a framework using large language models to predict molecular properties. The method leverages the ability of large language models to synthesize knowledge from literature and to reason about scientific data with domain expertise.

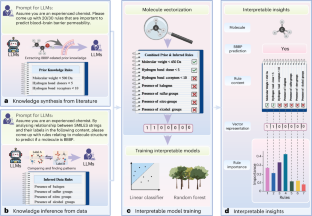
用于分子性质预测科学发现的大型语言模型
大型语言模型(llm)是人工智能系统的一种形式,它将大量的知识以自然语言的形式封装起来。这些系统擅长于许多复杂的任务,包括创意写作、讲故事、翻译、问答、摘要和计算机代码生成。尽管法学硕士在自然科学领域已经有了初步的应用,但它们推动科学发现的潜力在很大程度上仍未得到开发。在这项工作中,我们介绍了LLM4SD,这是一个框架,旨在利用llm通过综合文献知识和从科学数据中推断知识来推动分子性质预测的科学发现。法学硕士通过从科学文献中提取已建立的信息来合成知识,例如分子量是预测溶解度的关键。为了进行推理,llm识别分子数据中的模式,特别是在简化分子输入线输入系统编码的结构中,例如含卤素分子更有可能穿过血脑屏障。这些信息被呈现为可解释的知识,使分子转化为特征向量。通过将这些特征与随机森林等可解释模型一起使用,LLM4SD可以在预测分子特性的一系列基准任务中超越当前的技术水平。我们预计它将提供可解释的和潜在的新见解,有助于分子性质预测的科学发现。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Machine Intelligence
Multiple-
CiteScore
36.90
自引率
2.10%
发文量
127
期刊介绍:
Nature Machine Intelligence is a distinguished publication that presents original research and reviews on various topics in machine learning, robotics, and AI. Our focus extends beyond these fields, exploring their profound impact on other scientific disciplines, as well as societal and industrial aspects. We recognize limitless possibilities wherein machine intelligence can augment human capabilities and knowledge in domains like scientific exploration, healthcare, medical diagnostics, and the creation of safe and sustainable cities, transportation, and agriculture. Simultaneously, we acknowledge the emergence of ethical, social, and legal concerns due to the rapid pace of advancements.
To foster interdisciplinary discussions on these far-reaching implications, Nature Machine Intelligence serves as a platform for dialogue facilitated through Comments, News Features, News & Views articles, and Correspondence. Our goal is to encourage a comprehensive examination of these subjects.
Similar to all Nature-branded journals, Nature Machine Intelligence operates under the guidance of a team of skilled editors. We adhere to a fair and rigorous peer-review process, ensuring high standards of copy-editing and production, swift publication, and editorial independence.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


