Type III CRISPR-mediated flexible RNA excision with engineered guide RNAs
IF 14.5
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
Current RNA editing techniques are predominantly limited to single-base edits. Here, we introduce selective cleavages and intramolecular stitches of RNA (SCISSOR) for selective cleavage and intramolecular stitches of RNA. Building on the principle that type III CRISPR complex determines target cleavage positions based on gRNA length in 6-nt increments, we hypothesized that engineering gRNAs with bulge loops could circumvent this rule, allowing for flexible RNA excision. Through systematic evaluation of gRNAs with various bulge loops, we established the rules for precise non-6-nt target cleavage and repair. We observed that the complex tolerates 1- or 2-nt bulge loops and accommodates large bulge loops ranging from 6 to 24 nt. Consequently, SCISSOR could accomplish nearly any length of short fragment excision. With its capability to modify open reading frames, we demonstrate the potential of SCISSOR in repairing frameshift mutations and introducing frameshifts to create immunogenic poly-epitopes in human cells. SCISSOR holds promise in RNA therapy and biomedical research.
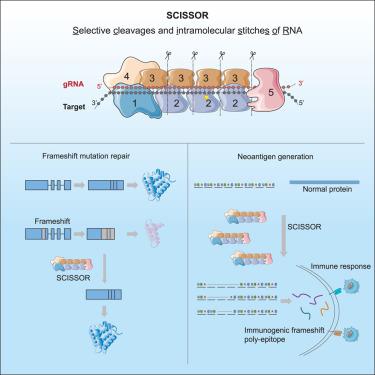
目前的 RNA 编辑技术主要局限于单碱基编辑。在这里,我们引入了选择性裂解和 RNA 分子内缝合(SCISSOR)技术,用于选择性裂解和 RNA 分子内缝合。基于 III 型 CRISPR 复合物根据 gRNA 长度以 6-nt 为增量确定目标切割位置的原理,我们假设具有隆起环的 gRNA 工程可以规避这一规则,从而实现灵活的 RNA 切除。通过对具有各种凸环的 gRNA 进行系统评估,我们建立了非 6-nt 目标精确切割和修复的规则。我们观察到,该复合体可容忍 1-nt 或 2-nt 的突起环,并可容纳 6 到 24 nt 的大突起环。因此,SCISSOR 几乎可以完成任何长度的短片段切割。凭借修改开放阅读框的能力,我们证明了 SCISSOR 在修复移帧突变和引入移帧以在人体细胞中创建免疫原性多表位方面的潜力。SCISSOR 为 RNA 治疗和生物医学研究带来了希望。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Molecular Cell
生物-生化与分子生物学
CiteScore
26.00
自引率
3.80%
发文量
389
审稿时长
1 months
期刊介绍:
Molecular Cell is a companion to Cell, the leading journal of biology and the highest-impact journal in the world. Launched in December 1997 and published monthly. Molecular Cell is dedicated to publishing cutting-edge research in molecular biology, focusing on fundamental cellular processes. The journal encompasses a wide range of topics, including DNA replication, recombination, and repair; Chromatin biology and genome organization; Transcription; RNA processing and decay; Non-coding RNA function; Translation; Protein folding, modification, and quality control; Signal transduction pathways; Cell cycle and checkpoints; Cell death; Autophagy; Metabolism.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


