Integrative spatial analysis reveals tumor heterogeneity and immune colony niche related to clinical outcomes in small cell lung cancer
IF 48.8
1区 医学
Q1 CELL BIOLOGY
引用次数: 0
Abstract
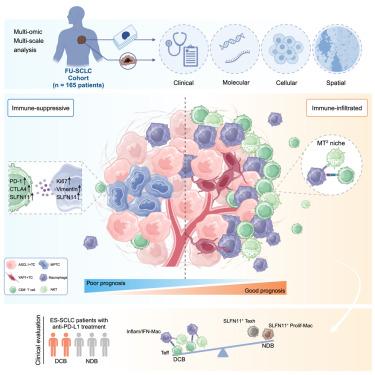
Recent advances have shed light on the molecular heterogeneity of small cell lung cancer (SCLC), yet the spatial organizations and cellular interactions in tumor immune microenvironment remain to be elucidated. Here, we employ co-detection by indexing (CODEX) and multi-omics profiling to delineate the spatial landscape for 165 SCLC patients, generating 267 high-dimensional images encompassing over 9.3 million cells. Integrating CODEX and genomic data reveals a multi-positive tumor cell neighborhood within ASCL1+ (SCLC-A) subtype, characterized by high SLFN11 expression and associated with poor prognosis. We further develop a cell colony detection algorithm (ColonyMap) and reveal a spatially assembled immune niche consisting of antitumoral macrophages, CD8+ T cells and natural killer T cells (MT2) which highly correlates with superior survival and predicts improving immunotherapy response in an independent cohort. This study serves as a valuable resource to study SCLC spatial heterogeneity and offers insights into potential patient stratification and personalized treatments.
求助全文
约1分钟内获得全文
求助全文
来源期刊

Cancer Cell
医学-肿瘤学
CiteScore
55.20
自引率
1.20%
发文量
179
审稿时长
4-8 weeks
期刊介绍:
Cancer Cell is a journal that focuses on promoting major advances in cancer research and oncology. The primary criteria for considering manuscripts are as follows:
Major advances: Manuscripts should provide significant advancements in answering important questions related to naturally occurring cancers.
Translational research: The journal welcomes translational research, which involves the application of basic scientific findings to human health and clinical practice.
Clinical investigations: Cancer Cell is interested in publishing clinical investigations that contribute to establishing new paradigms in the treatment, diagnosis, or prevention of cancers.
Insights into cancer biology: The journal values clinical investigations that provide important insights into cancer biology beyond what has been revealed by preclinical studies.
Mechanism-based proof-of-principle studies: Cancer Cell encourages the publication of mechanism-based proof-of-principle clinical studies, which demonstrate the feasibility of a specific therapeutic approach or diagnostic test.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


