Construction of a CRISPR-Cas9-Based Genetic Editing Tool for Serratia marcescens Using a Stationary Phase Promoter and Its Application in Putrescine Production
Abstract
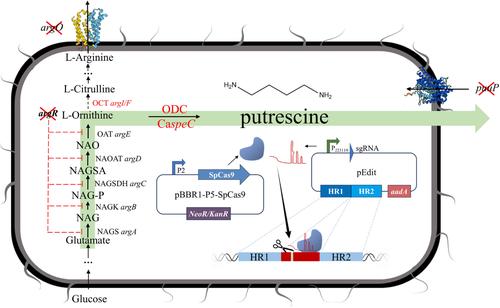
Putrescine plays a significant role in green food production and agriculture by promoting plant growth and enhancing crop quality. Its application reduces the reliance on chemical fertilizers and pesticides, thereby supporting the advancement of sustainable agricultural practices. This study achieved efficient production of putrescine in Serratia marcescens. S. marcescens has been extensively used to synthesize antimicrobial substances and express proteins, but its application has been limited by the lack of efficient genome-editing tools. This study presents a CRISPR-Cas9-based tool for gene editing in S. marcescens. A dual-plasmid system was constructed, incorporating an editing template into the plasmid pEdit with target-specific sgRNA. A stationary phase promoter was used to express Cas9 from Streptococcus pyogenes protein, avoiding the need for additional inducers and ensuring efficient one-step gene knockout and integration. The tool demonstrated over 80% editing efficiency across various S. marcescens strains and enabled successful single-base mutations. Using this tool, we enhanced putrescine production in S. marcescens HBQA7, optimizing the expression of ornithine decarboxylase from Clostridium aceticum DSM1496 with the P2 promoter and identifying the optimal integration site. Putrescine production reached 8.46 g/L within 48 h. This study significantly advances S. marcescens gene editing and metabolic engineering.


 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


