A unified cross-attention model for predicting antigen binding specificity to both HLA and TCR molecules
IF 18.8
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
Abstract
The immune checkpoint inhibitors have demonstrated promising clinical efficacy across various tumour types, yet the percentage of patients who benefit from them remains low. The bindings between tumour antigens and human leukocyte antigen class I/T cell receptor molecules determine the antigen presentation and T cell activation, thereby playing an important role in the immunotherapy response. In this paper, we propose UnifyImmun, a unified cross-attention transformer model designed to simultaneously predict the bindings of peptides to both receptors, providing more comprehensive evaluation of antigen immunogenicity. We devise a two-phase strategy using virtual adversarial training that enables these two tasks to reinforce each other mutually, by compelling the encoders to extract more expressive features. Our method demonstrates superior performance in predicting both peptide-HLA and peptide-TCR binding on multiple independent and external test sets. Notably, on a large-scale COVID-19 peptide-TCR binding test set without any seen peptide in the training set, our method outperforms the current state-of-the-art methods by more than 10%. The predicted binding scores significantly correlate with the immunotherapy response and clinical outcomes on two clinical cohorts. Furthermore, the cross-attention scores and integrated gradients reveal the amino acid sites critical for peptide binding to receptors. In essence, our approach marks an essential step towards comprehensive evaluation of antigen immunogenicity. This work proposes a deep learning model based on the cross-attention mechanism to simultaneously predict peptide–HLA and peptide–TCR bindings. Experiments verify that its performance for both prediction tasks on multiple test sets compares favourably with previous methods.

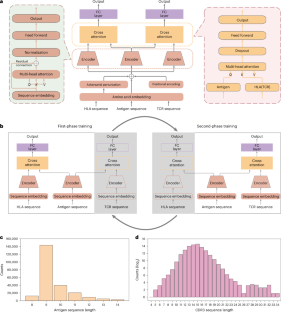
预测抗原结合HLA和TCR分子特异性的统一交叉注意模型
免疫检查点抑制剂在各种肿瘤类型中显示出有希望的临床疗效,但从中受益的患者比例仍然很低。肿瘤抗原与人白细胞抗原类I/T细胞受体分子的结合决定了抗原的呈递和T细胞的活化,从而在免疫治疗应答中发挥重要作用。在本文中,我们提出了统一的交叉注意转换模型UnifyImmun,旨在同时预测肽与两种受体的结合,提供更全面的抗原免疫原性评估。我们设计了一个使用虚拟对抗训练的两阶段策略,通过迫使编码器提取更多的表达特征,使这两个任务能够相互加强。我们的方法在预测多肽- hla和多肽- tcr结合的多个独立和外部测试集上表现出优异的性能。值得注意的是,在训练集中没有任何可见肽的大规模COVID-19肽- tcr结合测试集上,我们的方法比目前最先进的方法高出10%以上。在两个临床队列中,预测的结合评分与免疫治疗反应和临床结果显著相关。此外,交叉注意评分和综合梯度揭示了肽与受体结合的关键氨基酸位点。从本质上讲,我们的方法标志着抗原免疫原性综合评价的重要一步。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Machine Intelligence
Multiple-
CiteScore
36.90
自引率
2.10%
发文量
127
期刊介绍:
Nature Machine Intelligence is a distinguished publication that presents original research and reviews on various topics in machine learning, robotics, and AI. Our focus extends beyond these fields, exploring their profound impact on other scientific disciplines, as well as societal and industrial aspects. We recognize limitless possibilities wherein machine intelligence can augment human capabilities and knowledge in domains like scientific exploration, healthcare, medical diagnostics, and the creation of safe and sustainable cities, transportation, and agriculture. Simultaneously, we acknowledge the emergence of ethical, social, and legal concerns due to the rapid pace of advancements.
To foster interdisciplinary discussions on these far-reaching implications, Nature Machine Intelligence serves as a platform for dialogue facilitated through Comments, News Features, News & Views articles, and Correspondence. Our goal is to encourage a comprehensive examination of these subjects.
Similar to all Nature-branded journals, Nature Machine Intelligence operates under the guidance of a team of skilled editors. We adhere to a fair and rigorous peer-review process, ensuring high standards of copy-editing and production, swift publication, and editorial independence.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


