Liver fibrosis classification on trichrome histology slides using weakly supervised learning in children and young adults
Q2 Medicine
引用次数: 0
Abstract
Background
Traditional liver fibrosis staging via percutaneous biopsy suffers from sampling bias and variable inter-pathologist agreement, highlighting the need for more objective techniques. Deep learning models for disease staging from medical images have shown potential to decrease diagnostic variability, with recent weakly supervised learning strategies showing promising results even with limited manual annotation.
Purpose
To study the clustering-constrained attention multiple instance learning (CLAM) approach for staging liver fibrosis on trichrome whole slide images (WSIs) of children and young adults.
Methods
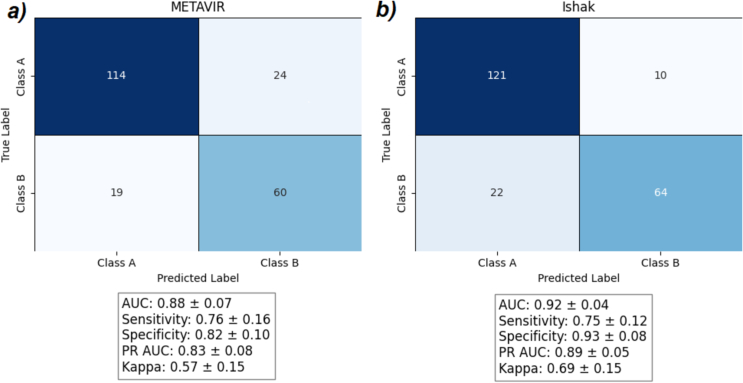
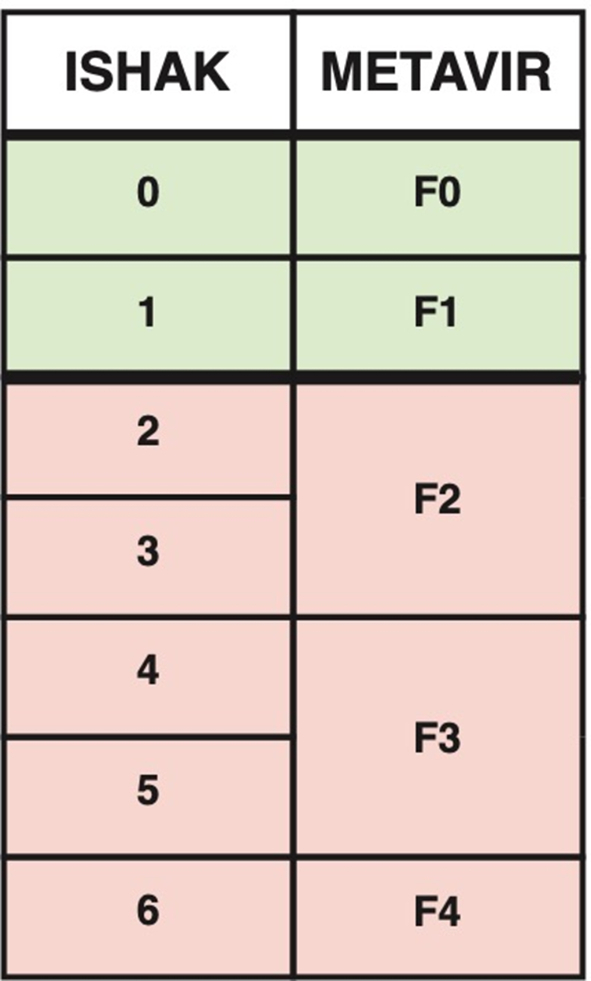
This is an ethics board approved retrospective study utilizing 217 trichrome WSI from pediatric liver biopsies for model development and testing. Two pediatric pathologists scored WSI using two liver fibrosis staging systems, METAVIR and Ishak. Cases were then secondarily categorized into either high- or low-stage liver fibrosis and used for model development. The CLAM pipeline was used to develop binary classification models for histological liver fibrosis. Model performance was evaluated using area under the curve (AUC), accuracy, sensitivity, specificity, and Cohen's Kappa.
Results
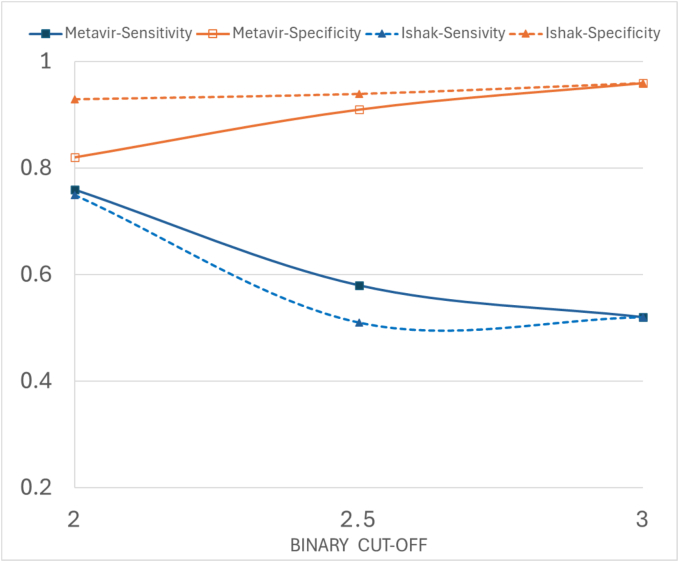
The CLAM models showed strong diagnostic performance, with sensitivities up to 0.76 and AUCs up to 0.92 for distinguishing low- and high-stage fibrosis. The agreement between model predictions and average pathologist scores was moderate to substantial (Kappa: 0.57–0.69), whereas pathologist agreement on the METAVIR and Ishak scoring systems was only fair (Kappa: 0.39–0.46).
Conclusions
CLAM pipeline showed promise in detecting features important for differentiating low- and high-stage fibrosis from trichrome WSI based on the results, offering a promising objective method for liver fibrosis detection in children and young adults.
在儿童和年轻人中使用弱监督学习的三色组织学切片的肝纤维化分类。
背景:传统的经皮活检肝纤维化分期存在抽样偏差和不同病理学家之间的一致性,强调需要更客观的技术。从医学图像中提取疾病分期的深度学习模型已经显示出降低诊断可变性的潜力,最近的弱监督学习策略即使在有限的人工注释下也显示出有希望的结果。目的:研究聚类约束注意多实例学习(CLAM)方法在儿童和青少年三色全切片(WSIs)肝纤维化分期中的应用。方法:这是一项伦理委员会批准的回顾性研究,利用217例小儿肝脏活检的三色WSI进行模型开发和测试。两名儿科病理学家使用METAVIR和Ishak两种肝纤维化分期系统对WSI进行评分。然后将病例二次分类为高或低阶段肝纤维化,并用于模型开发。CLAM管道用于建立组织学肝纤维化的二元分类模型。使用曲线下面积(AUC)、准确性、敏感性、特异性和Cohen’s Kappa来评估模型的性能。结果:CLAM模型显示出较强的诊断性能,在区分低分期和高分期纤维化方面的敏感性高达0.76,auc高达0.92。模型预测和平均病理学评分之间的一致性是中等到实质性的(Kappa: 0.57-0.69),而病理学家对METAVIR和Ishak评分系统的一致性只是公平的(Kappa: 0.39-0.46)。结论:基于结果,CLAM管道在检测三色WSI中区分低、高阶段纤维化的重要特征方面显示出前景,为儿童和年轻人的肝纤维化检测提供了一种有希望的客观方法。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Pathology Informatics
Medicine-Pathology and Forensic Medicine
CiteScore
3.70
自引率
0.00%
发文量
2
审稿时长
18 weeks
期刊介绍:
The Journal of Pathology Informatics (JPI) is an open access peer-reviewed journal dedicated to the advancement of pathology informatics. This is the official journal of the Association for Pathology Informatics (API). The journal aims to publish broadly about pathology informatics and freely disseminate all articles worldwide. This journal is of interest to pathologists, informaticians, academics, researchers, health IT specialists, information officers, IT staff, vendors, and anyone with an interest in informatics. We encourage submissions from anyone with an interest in the field of pathology informatics. We publish all types of papers related to pathology informatics including original research articles, technical notes, reviews, viewpoints, commentaries, editorials, symposia, meeting abstracts, book reviews, and correspondence to the editors. All submissions are subject to rigorous peer review by the well-regarded editorial board and by expert referees in appropriate specialties.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


