Kdm2a inhibition in skeletal muscle improves metabolic flexibility in obesity
IF 18.9
1区 医学
Q1 ENDOCRINOLOGY & METABOLISM
引用次数: 0
Abstract
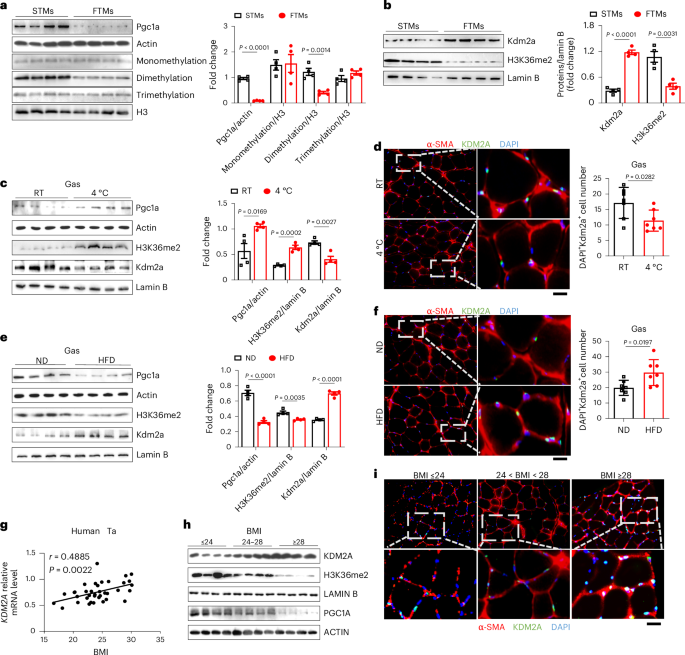
Skeletal muscle is a critical organ in maintaining homoeostasis against metabolic stress, and histone post-translational modifications are pivotal in those processes. However, the intricate nature of histone methylation in skeletal muscle and its impact on metabolic homoeostasis have yet to be elucidated. Here, we report that mitochondria-rich slow-twitch myofibers are characterized by significantly higher levels of H3K36me2 along with repressed expression of Kdm2a, an enzyme that specifically catalyses H3K36me2 demethylation. Deletion or inhibition of Kdm2a shifts fuel use from glucose under cold challenge to lipids under obese conditions by increasing the proportion of mitochondria-rich slow-twitch myofibers. This protects mice against cold insults and high-fat-diet-induced obesity and insulin resistance. Mechanistically, Kdm2a deficiency leads to a marked increase in H3K36me2 levels, which then promotes the recruitment of Mrg15 to the Esrrg locus to process its precursor messenger RNA splicing, thereby reshaping skeletal muscle metabolic profiles to induce slow-twitch myofiber transition. Collectively, our data support the role of Kdm2a as a viable target against metabolic stress. Kdm2a demethylates H3K36me2 to mediate fast-to-slow-twitch myofiber transition and reshape skeletal muscle metabolic flexibility under metabolic stress conditions.

抑制骨骼肌中的Kdm2a可改善肥胖患者的代谢灵活性
骨骼肌是维持体内平衡对抗代谢应激的关键器官,组蛋白翻译后修饰在这些过程中起关键作用。然而,骨骼肌中组蛋白甲基化的复杂性质及其对代谢平衡的影响尚未阐明。在这里,我们报告了富含线粒体的慢肌纤维的特征是H3K36me2水平显著升高,同时Kdm2a的表达受到抑制,Kdm2a是一种特异性催化H3K36me2去甲基化的酶。Kdm2a的缺失或抑制通过增加富含线粒体的慢肌纤维的比例,将燃料使用从寒冷挑战下的葡萄糖转移到肥胖条件下的脂质。这可以保护小鼠免受寒冷的伤害和高脂肪饮食引起的肥胖和胰岛素抵抗。从机制上说,Kdm2a缺乏导致H3K36me2水平显著增加,进而促进Mrg15募集到Esrrg位点,处理其前体信使RNA剪接,从而重塑骨骼肌代谢谱,诱导慢肌纤维转变。总的来说,我们的数据支持Kdm2a作为抗代谢应激的可行靶点的作用。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Nature metabolism
ENDOCRINOLOGY & METABOLISM-
CiteScore
27.50
自引率
2.40%
发文量
170
期刊介绍:
Nature Metabolism is a peer-reviewed scientific journal that covers a broad range of topics in metabolism research. It aims to advance the understanding of metabolic and homeostatic processes at a cellular and physiological level. The journal publishes research from various fields, including fundamental cell biology, basic biomedical and translational research, and integrative physiology. It focuses on how cellular metabolism affects cellular function, the physiology and homeostasis of organs and tissues, and the regulation of organismal energy homeostasis. It also investigates the molecular pathophysiology of metabolic diseases such as diabetes and obesity, as well as their treatment. Nature Metabolism follows the standards of other Nature-branded journals, with a dedicated team of professional editors, rigorous peer-review process, high standards of copy-editing and production, swift publication, and editorial independence. The journal has a high impact factor, has a certain influence in the international area, and is deeply concerned and cited by the majority of scholars.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


