Binding Selectivity of Inhibitors to BRD2 Uncovered by Molecular Docking and Molecular Dynamics Simulations
IF 2.9
4区 工程技术
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
Abstract
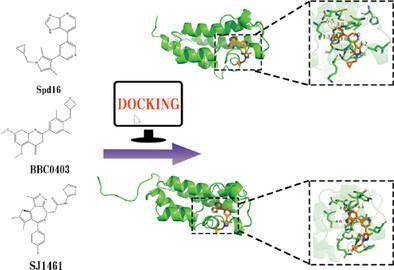
Bromodomain-containing protein 2 (BRD2) plays a significant role in the development and progression of various diseases. Investigating the binding selectivity of the two bromodomains BD1 and BD2 of BRD2 (BRD2-BD1 and BRD2-BD2) with specific inhibitors can provide valuable insights for rational design of novel therapeutic drugs. Thus, molecular dynamics (MD) simulations are employed to evaluate the selective mechanisms of three BRD2 inhibitors, Spd16, BBC0403, and SJ1461, toward BRD2-BD1 and BRD2-BD2. Molecular Mechanics/Generalized Born Surface Area (MM-GBSA) and Solvation Interaction Energy (SIE) methods are further employed to analyze the interaction modes and compare the binding free energies of BRD2-BD1 and BRD2-BD2 with these inhibitors. Although these inhibitors exert different effects on the internal structures of BRD2-BD1 and BRD2-BD2, their interaction patterns with these two bromodomains are similar. Phe372, Leu381, Tyr386, and Cys425 play key roles in the interaction between BRD2-BD2 and these three inhibitors, while Pro98, Leu108, Asn156, and Ile162 are the critical residues for the binding of BRD2-BD1 with these inhibitors. Non-polar interactions, particularly van der Waals interactions, serve as the primary driving force behind the binding of these inhibitors with BRD2-BD1 and BRD2-BD2. The findings of this study provide valuable insights for the rational design of novel BRD2 inhibitors.
求助全文
约1分钟内获得全文
求助全文
来源期刊

Advanced Theory and Simulations
Multidisciplinary-Multidisciplinary
CiteScore
5.50
自引率
3.00%
发文量
221
期刊介绍:
Advanced Theory and Simulations is an interdisciplinary, international, English-language journal that publishes high-quality scientific results focusing on the development and application of theoretical methods, modeling and simulation approaches in all natural science and medicine areas, including:
materials, chemistry, condensed matter physics
engineering, energy
life science, biology, medicine
atmospheric/environmental science, climate science
planetary science, astronomy, cosmology
method development, numerical methods, statistics
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


