Moving towards genome-wide data integration for patient stratification with Integrate Any Omics
IF 18.8
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
Abstract
High-throughput omics profiling advancements have greatly enhanced cancer patient stratification. However, incomplete data in multi-omics integration present a substantial challenge, as traditional methods like sample exclusion or imputation often compromise biological diversity and dependencies. Furthermore, the critical task of accurately classifying new patients with partial omics data into existing subtypes is commonly overlooked. To address these issues, we introduce Integrate Any Omics (IntegrAO), an unsupervised framework for integrating incomplete multi-omics data and classifying new samples. IntegrAO first combines partially overlapping patient graphs from diverse omics sources and utilizes graph neural networks to produce unified patient embeddings. Our systematic evaluation across five cancer cohorts involving six omics modalities demonstrates IntegrAO’s robustness to missing data and its accuracy in classifying new samples with partial profiles. An acute myeloid leukaemia case study further validates its capability to uncover biological and clinical heterogeneities in incomplete datasets. IntegrAO’s ability to handle heterogeneous and incomplete data makes it an essential tool for precision oncology, offering a holistic approach to patient characterization. Integrating incomplete multi-omics data remains a key challenge in precision oncology. IntegrAO, an unsupervised framework that integrates diverse omics, enables accurate patient classification even with incomplete datasets.

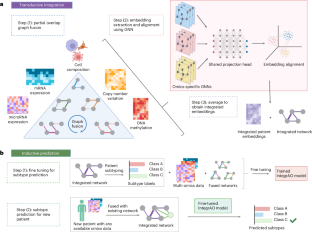
通过整合任意组学,向患者分层的全基因组数据整合迈进
高通量组学分析的进步极大地增强了癌症患者的分层。然而,在多组学整合中,数据的不完整带来了巨大的挑战,因为传统的方法如样本排除或imputation往往会损害生物多样性和依赖性。此外,将具有部分组学数据的新患者准确分类为现有亚型的关键任务通常被忽视。为了解决这些问题,我们引入了集成任意组学(IntegrAO),这是一个用于集成不完整多组学数据和分类新样本的无监督框架。IntegrAO首先将来自不同组学来源的部分重叠的患者图组合在一起,并利用图神经网络生成统一的患者嵌入。我们对涉及六种组学模式的五个癌症队列进行了系统评估,证明了IntegrAO对缺失数据的鲁棒性及其在分类具有部分特征的新样本方面的准确性。急性髓性白血病病例研究进一步验证了其在不完整数据集中揭示生物学和临床异质性的能力。IntegrAO处理异构和不完整数据的能力使其成为精确肿瘤学的重要工具,为患者表征提供了整体方法。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Machine Intelligence
Multiple-
CiteScore
36.90
自引率
2.10%
发文量
127
期刊介绍:
Nature Machine Intelligence is a distinguished publication that presents original research and reviews on various topics in machine learning, robotics, and AI. Our focus extends beyond these fields, exploring their profound impact on other scientific disciplines, as well as societal and industrial aspects. We recognize limitless possibilities wherein machine intelligence can augment human capabilities and knowledge in domains like scientific exploration, healthcare, medical diagnostics, and the creation of safe and sustainable cities, transportation, and agriculture. Simultaneously, we acknowledge the emergence of ethical, social, and legal concerns due to the rapid pace of advancements.
To foster interdisciplinary discussions on these far-reaching implications, Nature Machine Intelligence serves as a platform for dialogue facilitated through Comments, News Features, News & Views articles, and Correspondence. Our goal is to encourage a comprehensive examination of these subjects.
Similar to all Nature-branded journals, Nature Machine Intelligence operates under the guidance of a team of skilled editors. We adhere to a fair and rigorous peer-review process, ensuring high standards of copy-editing and production, swift publication, and editorial independence.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


