Genome-Scale Community Model-Guided Development of Bacterial Coculture for Lignocellulose Bioconversion
IF 3.5
2区 生物学
Q2 BIOTECHNOLOGY & APPLIED MICROBIOLOGY
引用次数: 0
Abstract
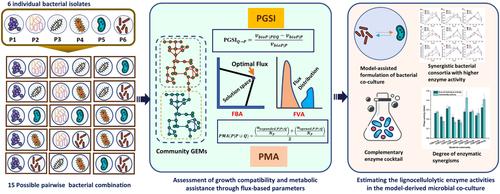
Microbial communities have shown promising potential in degrading complex biopolymers, producing value-added products through collaborative metabolic functionality. Hence, developing synthetic microbial consortia has become a predominant technique for various biotechnological applications. However, diverse microbial entities in a consortium can engage in distinct biochemical interactions that pose challenges in developing mutualistic communities. Therefore, a systems-level understanding of the inter-microbial metabolic interactions, growth compatibility, and metabolic synergisms is essential for developing effective synthetic consortia. This study demonstrated a genome-scale community modeling approach to assess the inter-microbial interaction pattern and screen metabolically compatible bacterial pairs for designing the lignocellulolytic coculture system. Here, we have investigated the pairwise growth and biochemical synergisms among six termite gut bacterial isolates by implementing flux-based parameters, i.e., pairwise growth support index (PGSI) and metabolic assistance (PMA). Assessment of the PGSI and PMA helps screen nine beneficial bacterial pairs that were validated by designing a coculture experiment with lignocellulosic substrates. For the cocultured bacterial pairs, the experimentally measured enzymatic synergisms (DES) showed good coherence with model-derived biochemical compatibility (PMA), which explains the fidelity of the in silico predictions. The highest degree of enzymatic synergisms has been observed in C. denverensis P3 and Brevibacterium sp P5 coculture, where the total cellulase activity has been increased by 53%. Hence, the flux-based assessment of inter-microbial interactions and metabolic compatibility helps select the best bacterial coculture system with enhanced lignocellulolytic functionality. The flux-based parameters (PGSI and PMA) in the proposed community modeling strategy will help optimize the composition of microbial consortia for developing synthetic microcosms for bioremediation, bioengineering, and biomedical applications.
求助全文
约1分钟内获得全文
求助全文
来源期刊

Biotechnology and Bioengineering
工程技术-生物工程与应用微生物
CiteScore
7.90
自引率
5.30%
发文量
280
审稿时长
2.1 months
期刊介绍:
Biotechnology & Bioengineering publishes Perspectives, Articles, Reviews, Mini-Reviews, and Communications to the Editor that embrace all aspects of biotechnology. These include:
-Enzyme systems and their applications, including enzyme reactors, purification, and applied aspects of protein engineering
-Animal-cell biotechnology, including media development
-Applied aspects of cellular physiology, metabolism, and energetics
-Biocatalysis and applied enzymology, including enzyme reactors, protein engineering, and nanobiotechnology
-Biothermodynamics
-Biofuels, including biomass and renewable resource engineering
-Biomaterials, including delivery systems and materials for tissue engineering
-Bioprocess engineering, including kinetics and modeling of biological systems, transport phenomena in bioreactors, bioreactor design, monitoring, and control
-Biosensors and instrumentation
-Computational and systems biology, including bioinformatics and genomic/proteomic studies
-Environmental biotechnology, including biofilms, algal systems, and bioremediation
-Metabolic and cellular engineering
-Plant-cell biotechnology
-Spectroscopic and other analytical techniques for biotechnological applications
-Synthetic biology
-Tissue engineering, stem-cell bioengineering, regenerative medicine, gene therapy and delivery systems
The editors will consider papers for publication based on novelty, their immediate or future impact on biotechnological processes, and their contribution to the advancement of biochemical engineering science. Submission of papers dealing with routine aspects of bioprocessing, description of established equipment, and routine applications of established methodologies (e.g., control strategies, modeling, experimental methods) is discouraged. Theoretical papers will be judged based on the novelty of the approach and their potential impact, or on their novel capability to predict and elucidate experimental observations.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


