Nucleosomal asymmetry shapes histone mark binding and promotes poising at bivalent domains
IF 14.5
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
Promoters of developmental genes in embryonic stem cells (ESCs) are marked by histone H3 lysine 4 trimethylation (H3K4me3) and H3K27me3 in an asymmetric nucleosomal conformation, with each sister histone H3 carrying only one of the two marks. These bivalent domains are thought to poise genes for timely activation upon differentiation. Here, we show that asymmetric bivalent nucleosomes recruit repressive H3K27me3 binders but fail to enrich activating H3K4me3 binders, thereby promoting a poised state. Strikingly, the bivalent mark combination further promotes recruitment of specific chromatin proteins that are not recruited by each mark individually, including the lysine acetyltransferase (KAT) complex KAT6B. Knockout of KAT6B blocks neuronal differentiation, demonstrating that KAT6B is critical for proper bivalent gene expression during ESC differentiation. These findings reveal how readout of the bivalent histone marks directly promotes a poised state at developmental genes while highlighting how nucleosomal asymmetry is critical for histone mark readout and function.
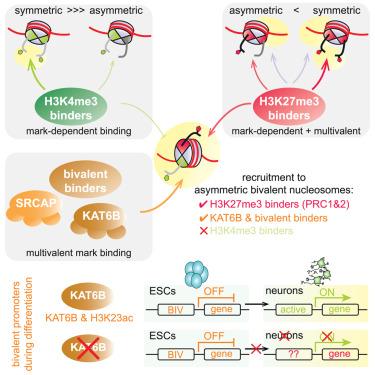
核小体不对称形成组蛋白标记结合并促进二价结构域的定位
胚胎干细胞(ESCs)中发育基因的启动子由组蛋白H3赖氨酸4三甲基化(H3K4me3)和H3K27me3以不对称核小体构象标记,每个姐妹组蛋白H3只携带两个标记中的一个。这些二价结构域被认为是平衡基因在分化时的及时激活。在这里,我们发现不对称二价核小体招募抑制H3K27me3结合物,但不能富集激活H3K4me3结合物,从而促进平衡状态。引人注目的是,二价标记组合进一步促进了特定染色质蛋白的募集,这些蛋白不是由每个标记单独募集的,包括赖氨酸乙酰转移酶(KAT)复合物KAT6B。敲除KAT6B可阻断神经元分化,表明KAT6B对ESC分化过程中二价基因的表达至关重要。这些发现揭示了二价组蛋白标记的读取如何直接促进发育基因的平衡状态,同时强调了核小体不对称如何对组蛋白标记的读取和功能至关重要。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Molecular Cell
生物-生化与分子生物学
CiteScore
26.00
自引率
3.80%
发文量
389
审稿时长
1 months
期刊介绍:
Molecular Cell is a companion to Cell, the leading journal of biology and the highest-impact journal in the world. Launched in December 1997 and published monthly. Molecular Cell is dedicated to publishing cutting-edge research in molecular biology, focusing on fundamental cellular processes. The journal encompasses a wide range of topics, including DNA replication, recombination, and repair; Chromatin biology and genome organization; Transcription; RNA processing and decay; Non-coding RNA function; Translation; Protein folding, modification, and quality control; Signal transduction pathways; Cell cycle and checkpoints; Cell death; Autophagy; Metabolism.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


