Formyl-methionine-mediated eukaryotic ribosome quality control pathway for cold adaptation
IF 16.6
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
Protein synthesis in the eukaryotic cytosol can start using both conventional methionine and formyl-methionine (fMet). However, a mechanism, if such exists, for detecting and regulating the incorporation of fMet (instead of Met) during translation, thereby preventing cellular toxicity of nascent fMet-bearing (fMet-) polypeptides, remains unknown. Here, we describe the fMet-mediated ribosome quality control (fMet-RQC) pathway in Saccharomyces cerevisiae. A eukaryotic translation initiation factor 3 subunit c, Nip1, specifically recognizes N-terminal fMet in nascent polypeptides, recruiting a small GTPase, Arf1, to induce ribosome stalling, largely with 41-residue fMet-peptidyl tRNAs. This leads to ribosome dissociation and subsequent stress granule formation. Loss of the fMet-RQC pathway causes the continued synthesis of fMet polypeptides, which inhibits essential N-terminal Met modifications and promotes their coaggregation with ribosomes. This fMet-RQC pathway is important for the adaptation of yeast cells to cold stress by promoting stress granule formation and preventing a buildup of toxic fMet polypeptides.
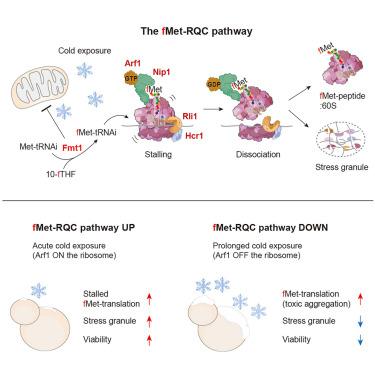
甲蛋氨酸介导的真核核糖体冷适应质量控制途径
真核细胞质中的蛋白质合成可以同时使用传统蛋氨酸和甲酰基蛋氨酸(fMet)。然而,一种机制,如果存在的话,检测和调节翻译过程中fMet(而不是Met)的结合,从而防止新生的含fMet (fMet-)多肽的细胞毒性,仍然未知。在这里,我们描述了酿酒酵母中fmet介导的核糖体质量控制(fMet-RQC)途径。真核生物翻译起始因子3亚基c Nip1特异性识别新生多肽中的n端fMet,招募一个小的GTPase Arf1诱导核糖体停滞,主要是41个残基fMet肽基trna。这导致核糖体解离和随后的应力颗粒形成。fMet- rqc通路的缺失会导致fMet多肽的持续合成,从而抑制必需的n端Met修饰并促进其与核糖体的共聚集。这种fMet- rqc途径通过促进应激颗粒的形成和防止有毒fMet多肽的积累,对酵母细胞适应冷胁迫很重要。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Molecular Cell
生物-生化与分子生物学
CiteScore
26.00
自引率
3.80%
发文量
389
审稿时长
1 months
期刊介绍:
Molecular Cell is a companion to Cell, the leading journal of biology and the highest-impact journal in the world. Launched in December 1997 and published monthly. Molecular Cell is dedicated to publishing cutting-edge research in molecular biology, focusing on fundamental cellular processes. The journal encompasses a wide range of topics, including DNA replication, recombination, and repair; Chromatin biology and genome organization; Transcription; RNA processing and decay; Non-coding RNA function; Translation; Protein folding, modification, and quality control; Signal transduction pathways; Cell cycle and checkpoints; Cell death; Autophagy; Metabolism.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


