Mutability and hypermutation antagonize immunoglobulin codon optimality
IF 14.5
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
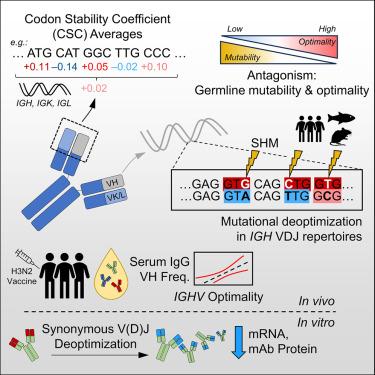
The efficacy of antibody responses is inherently linked to paratope diversity, as generated through V(D)J recombination and somatic hypermutation. Despite this, it is unclear how genetic diversification mechanisms evolved alongside codon optimality and affect antibody expression. Here, we analyze germline immunoglobulin (IG) genes, natural V(D)J repertoires, serum IgG, and monoclonal antibody (mAb) expression through the lens of codon optimality. Germline variable genes (IGVs) exhibit diverse optimality that is inversely related to mutability. Hypermutation deoptimizes heavy-chain (IGH) VDJ repertoires within human tonsils, bone marrow, lymph nodes (including SARS-CoV-2-specific clones), blood (HIV-1-specific clones), mice, and zebrafish. Analyses of mutation-affected codons show that targeting to complementarity-determining regions constrains deoptimization. Germline IGHV optimality correlates with serum variable fragment (VH) usage after influenza vaccination, while synonymous deoptimization attenuated mAb yield. These findings provide unanticipated insights into an antagonistic relationship between diversification mechanisms and codon optimality. Ultimately, the need for diversity takes precedence over that for the most optimal codon usage.
求助全文
约1分钟内获得全文
求助全文
来源期刊

Molecular Cell
生物-生化与分子生物学
CiteScore
26.00
自引率
3.80%
发文量
389
审稿时长
1 months
期刊介绍:
Molecular Cell is a companion to Cell, the leading journal of biology and the highest-impact journal in the world. Launched in December 1997 and published monthly. Molecular Cell is dedicated to publishing cutting-edge research in molecular biology, focusing on fundamental cellular processes. The journal encompasses a wide range of topics, including DNA replication, recombination, and repair; Chromatin biology and genome organization; Transcription; RNA processing and decay; Non-coding RNA function; Translation; Protein folding, modification, and quality control; Signal transduction pathways; Cell cycle and checkpoints; Cell death; Autophagy; Metabolism.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


