Leveraging deep learning for identification and segmentation of “CAF-1/p60-positive” nuclei in oral squamous cell carcinoma tissue samples
Q2 Medicine
引用次数: 0
Abstract
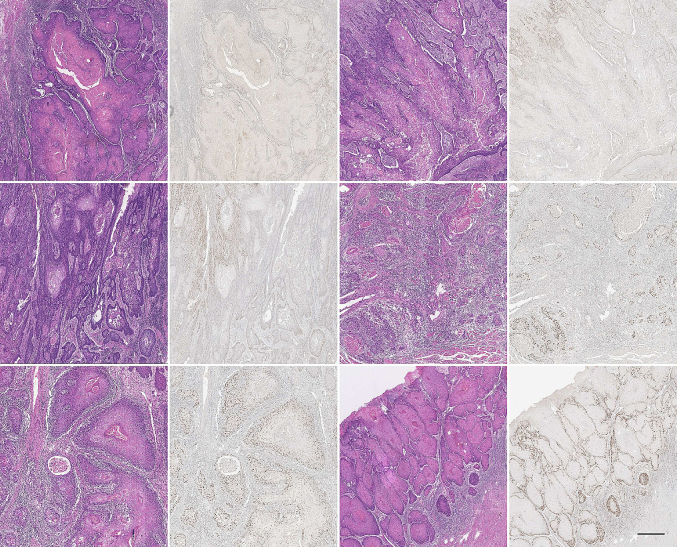
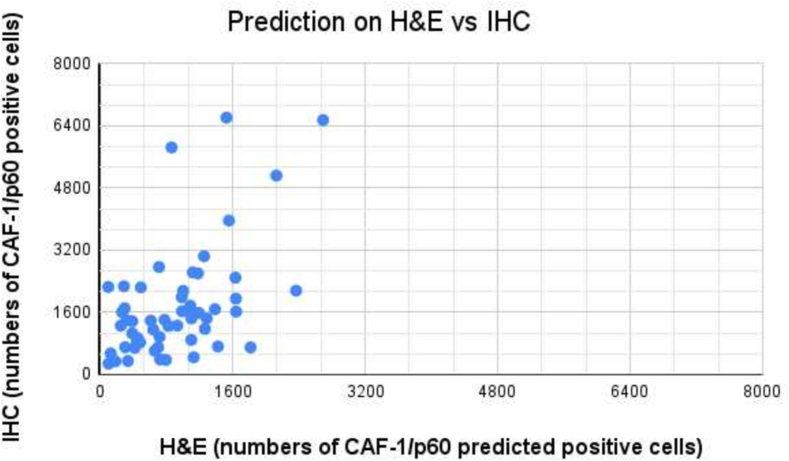
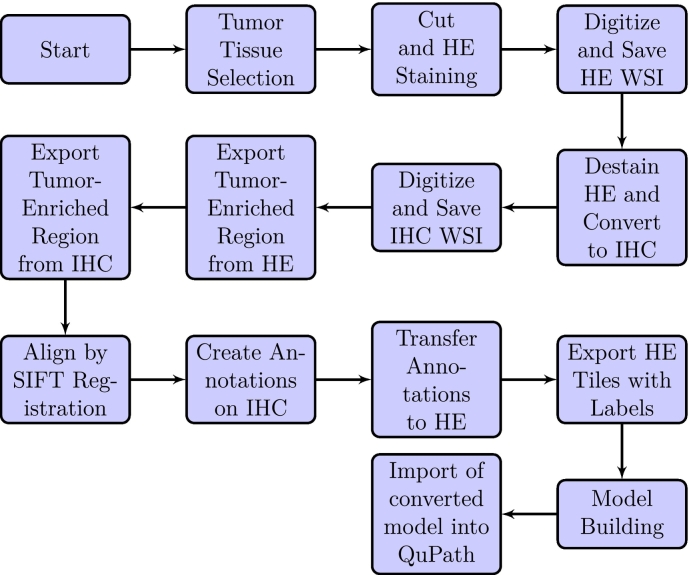
In the current study, we introduced a unique method for identifying and segmenting oral squamous cell carcinoma (OSCC) nuclei, concentrating on those predicted to have significant CAF-1/p60 protein expression. Our suggested model uses the StarDist architecture, a deep-learning framework designed for biomedical image segmentation tasks. The training dataset comprises painstakingly annotated masks created from tissue sections previously stained with hematoxylin and eosin (H&E) and then restained with immunohistochemistry (IHC) for p60 protein. Our algorithm uses subtle morphological and colorimetric H&E cellular characteristics to predict CAF-1/p60 IHC expression in OSCC nuclei. The StarDist-based architecture performs exceptionally well in localizing and segmenting H&E nuclei, previously identified by IHC-based ground truth. In summary, our innovative approach harnesses deep learning and multimodal information to advance the automated analysis of OSCC nuclei exhibiting specific protein expression patterns. This methodology holds promise for expediting accurate pathological assessment and gaining deeper insights into the role of CAF-1/p60 protein within the context of oral cancer progression.
利用深度学习在口腔鳞状细胞癌组织样本中识别和分割“ca -1/p60阳性”细胞核。
在目前的研究中,我们介绍了一种独特的方法来识别和分割口腔鳞状细胞癌(OSCC)细胞核,集中在那些预测有显著的ca -1/p60蛋白表达。我们建议的模型使用StarDist架构,这是一种为生物医学图像分割任务设计的深度学习框架。训练数据集包括精心注释的掩膜,这些掩膜是由先前用苏木精和伊红(H&E)染色的组织切片创建的,然后用免疫组织化学(IHC)保留p60蛋白。我们的算法使用细微的形态学和比色H&E细胞特征来预测ca -1/p60 IHC在OSCC细胞核中的表达。基于stardist的架构在定位和分割H&E核方面表现得非常好,以前是由基于ihc的ground truth识别的。总之,我们的创新方法利用深度学习和多模态信息来推进显示特定蛋白质表达模式的OSCC核的自动分析。这种方法有望加快准确的病理评估,并更深入地了解ca -1/p60蛋白在口腔癌进展中的作用。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Pathology Informatics
Medicine-Pathology and Forensic Medicine
CiteScore
3.70
自引率
0.00%
发文量
2
审稿时长
18 weeks
期刊介绍:
The Journal of Pathology Informatics (JPI) is an open access peer-reviewed journal dedicated to the advancement of pathology informatics. This is the official journal of the Association for Pathology Informatics (API). The journal aims to publish broadly about pathology informatics and freely disseminate all articles worldwide. This journal is of interest to pathologists, informaticians, academics, researchers, health IT specialists, information officers, IT staff, vendors, and anyone with an interest in informatics. We encourage submissions from anyone with an interest in the field of pathology informatics. We publish all types of papers related to pathology informatics including original research articles, technical notes, reviews, viewpoints, commentaries, editorials, symposia, meeting abstracts, book reviews, and correspondence to the editors. All submissions are subject to rigorous peer review by the well-regarded editorial board and by expert referees in appropriate specialties.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


