Leveraging ancestral sequence reconstruction for protein representation learning
IF 18.8
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
Abstract
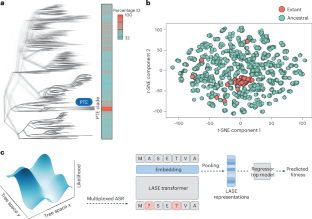
Protein language models (PLMs) convert amino acid sequences into the numerical representations required to train machine learning models. Many PLMs are large (>600 million parameters) and trained on a broad span of protein sequence space. However, these models have limitations in terms of predictive accuracy and computational cost. Here we use multiplexed ancestral sequence reconstruction to generate small but focused functional protein sequence datasets for PLM training. Compared to large PLMs, this local ancestral sequence embedding produces representations with higher predictive accuracy. We show that due to the evolutionary nature of the ancestral sequence reconstruction data, local ancestral sequence embedding produces smoother fitness landscapes, in which protein variants that are closer in fitness value become numerically closer in representation space. This work contributes to the implementation of machine learning-based protein design in real-world settings, where data are sparse and computational resources are limited. Matthews et al. present a protein sequence embedding based on data from ancestral sequences that allows machine learning to be used for tasks where training data are scarce or expensive.

利用祖先序列重建进行蛋白质表示学习
蛋白质语言模型(PLM)将氨基酸序列转换为训练机器学习模型所需的数字表示。许多蛋白质语言模型都很庞大(6 亿个参数),并在广泛的蛋白质序列空间中进行训练。然而,这些模型在预测准确性和计算成本方面存在局限性。在这里,我们使用多路复用祖先序列重建来生成小而集中的功能蛋白质序列数据集,用于PLM训练。与大型 PLM 相比,这种局部祖先序列嵌入产生的表征具有更高的预测准确性。我们的研究表明,由于祖先序列重构数据的进化性质,局部祖先序列嵌入会产生更平滑的适配性景观,在这种景观中,适配值更接近的蛋白质变体在表示空间的数值上也更接近。这项工作有助于在数据稀少、计算资源有限的实际环境中实现基于机器学习的蛋白质设计。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Machine Intelligence
Multiple-
CiteScore
36.90
自引率
2.10%
发文量
127
期刊介绍:
Nature Machine Intelligence is a distinguished publication that presents original research and reviews on various topics in machine learning, robotics, and AI. Our focus extends beyond these fields, exploring their profound impact on other scientific disciplines, as well as societal and industrial aspects. We recognize limitless possibilities wherein machine intelligence can augment human capabilities and knowledge in domains like scientific exploration, healthcare, medical diagnostics, and the creation of safe and sustainable cities, transportation, and agriculture. Simultaneously, we acknowledge the emergence of ethical, social, and legal concerns due to the rapid pace of advancements.
To foster interdisciplinary discussions on these far-reaching implications, Nature Machine Intelligence serves as a platform for dialogue facilitated through Comments, News Features, News & Views articles, and Correspondence. Our goal is to encourage a comprehensive examination of these subjects.
Similar to all Nature-branded journals, Nature Machine Intelligence operates under the guidance of a team of skilled editors. We adhere to a fair and rigorous peer-review process, ensuring high standards of copy-editing and production, swift publication, and editorial independence.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


