An interpretable RNA foundation model for exploring functional RNA motifs in plants
IF 18.8
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
Abstract
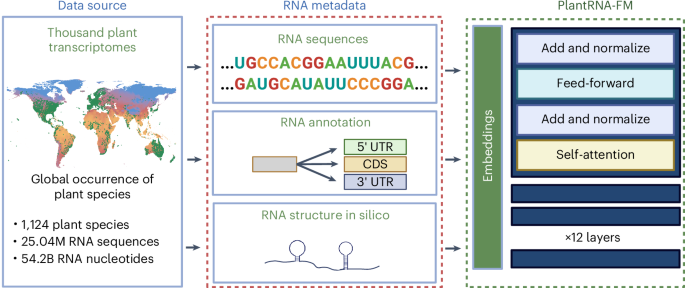
The complex ‘language’ of plant RNA encodes a vast array of biological regulatory elements that orchestrate crucial aspects of plant growth, development and adaptation to environmental stresses. Recent advancements in foundation models (FMs) have demonstrated their unprecedented potential to decipher complex ‘language’ in biology. In this study, we introduced PlantRNA-FM, a high-performance and interpretable RNA FM specifically designed for plants. PlantRNA-FM was pretrained on an extensive dataset, integrating RNA sequences and RNA structure information from 1,124 distinct plant species. PlantRNA-FM exhibits superior performance in plant-specific downstream tasks. PlantRNA-FM achieves an F1 score of 0.974 for genic region annotation, whereas the current best-performing model achieves 0.639. Our PlantRNA-FM is empowered by our interpretable framework that facilitates the identification of biologically functional RNA sequence and structure motifs, including both RNA secondary and tertiary structure motifs across transcriptomes. Through experimental validations, we revealed translation-associated RNA motifs in plants. Our PlantRNA-FM also highlighted the importance of the position information of these functional RNA motifs in genic regions. Taken together, our PlantRNA-FM facilitates the exploration of functional RNA motifs across the complexity of transcriptomes, empowering plant scientists with capabilities for programming RNA codes in plants. Approaches are needed to explore regulatory RNA motifs in plants. An interpretable RNA foundation model is developed, trained on thousands of plant transcriptomes, which achieves superior performance in plant RNA biology tasks and enables the discovery of functional RNA sequence and structure motifs across transcriptomes.

探索植物功能性RNA基序的可解释RNA基础模型
植物RNA的复杂“语言”编码了大量的生物调控元件,这些元件协调了植物生长、发育和适应环境胁迫的关键方面。基础模型(FMs)的最新进展显示了它们在破译生物学中复杂“语言”方面前所未有的潜力。在这项研究中,我们引入了PlantRNA-FM,一种专门为植物设计的高性能、可解释的RNA FM。PlantRNA-FM是在一个广泛的数据集上进行预训练的,该数据集整合了来自1,124种不同植物的RNA序列和RNA结构信息。PlantRNA-FM在植物特异性下游任务中表现出优越的性能。PlantRNA-FM对基因区域注释的F1得分为0.974,而目前表现最好的模型为0.639。我们的PlantRNA-FM由我们的可解释框架授权,有助于识别生物功能RNA序列和结构基序,包括转录组中的RNA二级和三级结构基序。通过实验验证,我们揭示了植物中翻译相关的RNA基序。我们的PlantRNA-FM也强调了这些功能RNA基序在基因区域的位置信息的重要性。综上所述,我们的PlantRNA-FM有助于在转录组的复杂性中探索功能性RNA基序,使植物科学家能够编程植物中的RNA编码。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Machine Intelligence
Multiple-
CiteScore
36.90
自引率
2.10%
发文量
127
期刊介绍:
Nature Machine Intelligence is a distinguished publication that presents original research and reviews on various topics in machine learning, robotics, and AI. Our focus extends beyond these fields, exploring their profound impact on other scientific disciplines, as well as societal and industrial aspects. We recognize limitless possibilities wherein machine intelligence can augment human capabilities and knowledge in domains like scientific exploration, healthcare, medical diagnostics, and the creation of safe and sustainable cities, transportation, and agriculture. Simultaneously, we acknowledge the emergence of ethical, social, and legal concerns due to the rapid pace of advancements.
To foster interdisciplinary discussions on these far-reaching implications, Nature Machine Intelligence serves as a platform for dialogue facilitated through Comments, News Features, News & Views articles, and Correspondence. Our goal is to encourage a comprehensive examination of these subjects.
Similar to all Nature-branded journals, Nature Machine Intelligence operates under the guidance of a team of skilled editors. We adhere to a fair and rigorous peer-review process, ensuring high standards of copy-editing and production, swift publication, and editorial independence.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


