Evaluating generalizability of artificial intelligence models for molecular datasets
IF 18.8
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
Abstract
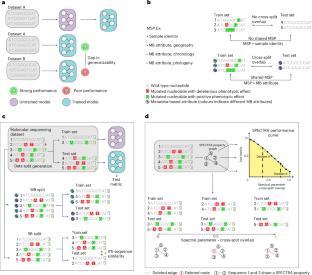
Deep learning has made rapid advances in modelling molecular sequencing data. Despite achieving high performance on benchmarks, it remains unclear to what extent deep learning models learn general principles and generalize to previously unseen sequences. Benchmarks traditionally interrogate model generalizability by generating metadata- or sequence similarity-based train and test splits of input data before assessing model performance. Here we show that this approach mischaracterizes model generalizability by failing to consider the full spectrum of cross-split overlap, that is, similarity between train and test splits. We introduce SPECTRA, the spectral framework for model evaluation. Given a model and a dataset, SPECTRA plots model performance as a function of decreasing cross-split overlap and reports the area under this curve as a measure of generalizability. We use SPECTRA with 18 sequencing datasets and phenotypes ranging from antibiotic resistance in tuberculosis to protein–ligand binding and evaluate the generalizability of 19 state-of-the-art deep learning models, including large language models, graph neural networks, diffusion models and convolutional neural networks. We show that sequence similarity- and metadata-based splits provide an incomplete assessment of model generalizability. Using SPECTRA, we find that as cross-split overlap decreases, deep learning models consistently show reduced performance, varying by task and model. Although no model consistently achieved the highest performance across all tasks, deep learning models can, in some cases, generalize to previously unseen sequences on specific tasks. SPECTRA advances our understanding of how foundation models generalize in biological applications. Ektefaie and colleagues introduce the spectral framework for models evaluation (SPECTRA) to measure the generalizability of machine learning models for molecular sequences.

评估人工智能模型在分子数据集上的泛化性
深度学习在分子测序数据建模方面取得了快速进展。尽管在基准测试中取得了很高的性能,但深度学习模型在多大程度上学习了一般原理,并将其推广到以前未见过的序列,这一点尚不清楚。在评估模型性能之前,基准测试通常通过生成基于元数据或序列相似性的训练和测试输入数据的分割来询问模型的泛化性。在这里,我们表明,这种方法错误地描述了模型的可泛化性,因为它没有考虑交叉分割重叠的全谱,即训练和测试分割之间的相似性。我们介绍了用于模型评估的光谱框架SPECTRA。给定一个模型和一个数据集,SPECTRA将模型性能绘制为减少交叉分裂重叠的函数,并报告该曲线下的面积作为泛化性的度量。我们将SPECTRA与18个测序数据集和表型(从结核病的抗生素耐药性到蛋白质配体结合)一起使用,并评估了19个最先进的深度学习模型的泛化性,包括大型语言模型、图神经网络、扩散模型和卷积神经网络。我们表明,序列相似性和基于元数据的分裂提供了模型泛化的不完全评估。使用SPECTRA,我们发现随着交叉分割重叠的减少,深度学习模型的性能持续下降,因任务和模型而异。虽然没有模型在所有任务中都能达到最高的性能,但在某些情况下,深度学习模型可以在特定任务中推广到以前未见过的序列。SPECTRA提高了我们对基础模型如何在生物学应用中推广的理解。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Machine Intelligence
Multiple-
CiteScore
36.90
自引率
2.10%
发文量
127
期刊介绍:
Nature Machine Intelligence is a distinguished publication that presents original research and reviews on various topics in machine learning, robotics, and AI. Our focus extends beyond these fields, exploring their profound impact on other scientific disciplines, as well as societal and industrial aspects. We recognize limitless possibilities wherein machine intelligence can augment human capabilities and knowledge in domains like scientific exploration, healthcare, medical diagnostics, and the creation of safe and sustainable cities, transportation, and agriculture. Simultaneously, we acknowledge the emergence of ethical, social, and legal concerns due to the rapid pace of advancements.
To foster interdisciplinary discussions on these far-reaching implications, Nature Machine Intelligence serves as a platform for dialogue facilitated through Comments, News Features, News & Views articles, and Correspondence. Our goal is to encourage a comprehensive examination of these subjects.
Similar to all Nature-branded journals, Nature Machine Intelligence operates under the guidance of a team of skilled editors. We adhere to a fair and rigorous peer-review process, ensuring high standards of copy-editing and production, swift publication, and editorial independence.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


