Nanobody–antigen interaction prediction with ensemble deep learning and prompt-based protein language models
IF 18.8
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
Abstract
Nanobodies can provide specific binding to divergent antigens, leading to many promising therapeutic and detection applications in recent years. Traditional technologies of nanobody discovery based on alpaca immunization and phage display are very time-consuming and labour-intensive. Despite recent progress in the study of nanobodies, developing fast and accurate computational tools for nanobody–antigen interaction (NAI) prediction is urgently desirable. Here we propose an ensemble deep learning-based framework named DeepNano-seq to predict general protein–protein interaction (PPI) containing NAI from pure sequence information. Quantitative comparison results show that DeepNano-seq possesses the best cross-species generalization ability among existing PPI algorithms. Nevertheless, several of the most effective PPI methods, including DeepNano-seq, demonstrate suboptimal performance for NAI prediction due to the distinction between NAI and PPI at both the pattern and data levels. Therefore, we organize NAI data from the public database for dedicated NAI modelling. Furthermore, we enhance the prediction pipeline of DeepNano-seq by directing the model’s attention to the antigen-binding sites through a prompt-based approach to present the final DeepNano. The comprehensive evaluation demonstrates that DeepNano performs superiorly in NAI prediction and virtual screening of nanobodies. Overall, DeepNano-seq and DeepNano can offer powerful tools for nanobody discovery. Predicting nanobody–antigen interactions is crucial for advancing nanobody development in drug discovery, but it remains a challenging task. Deng et al. propose DeepNano to enhance the prediction of nanobody–antigen interactions, facilitating virtual screening of target nanobodies.
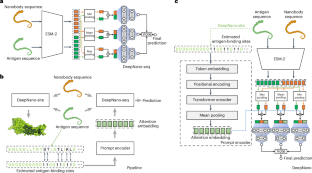

纳米体-抗原相互作用预测与集成深度学习和基于提示的蛋白质语言模型
纳米体可以提供特异性结合抗原,导致近年来许多有前景的治疗和检测应用。传统的基于羊驼免疫和噬菌体展示的纳米体发现技术非常耗时和费力。尽管近年来纳米体的研究取得了进展,但迫切需要开发快速准确的计算工具来预测纳米体-抗原相互作用(NAI)。在这里,我们提出了一个基于集成深度学习的框架,名为DeepNano-seq,用于从纯序列信息中预测含有NAI的一般蛋白质-蛋白质相互作用(PPI)。定量比较结果表明,在现有的PPI算法中,DeepNano-seq具有最好的跨物种泛化能力。然而,包括DeepNano-seq在内的几种最有效的PPI方法,由于NAI和PPI在模式和数据水平上的差异,在NAI预测中表现出了次优的性能。因此,我们从公共数据库中组织NAI数据,用于专门的NAI建模。此外,我们通过基于提示的方法将模型的注意力引导到抗原结合位点,从而增强了DeepNano-seq的预测管道。综合评价表明,DeepNano在纳米体的NAI预测和虚拟筛选方面表现优异。总之,DeepNano-seq和DeepNano可以为纳米体的发现提供强大的工具。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Machine Intelligence
Multiple-
CiteScore
36.90
自引率
2.10%
发文量
127
期刊介绍:
Nature Machine Intelligence is a distinguished publication that presents original research and reviews on various topics in machine learning, robotics, and AI. Our focus extends beyond these fields, exploring their profound impact on other scientific disciplines, as well as societal and industrial aspects. We recognize limitless possibilities wherein machine intelligence can augment human capabilities and knowledge in domains like scientific exploration, healthcare, medical diagnostics, and the creation of safe and sustainable cities, transportation, and agriculture. Simultaneously, we acknowledge the emergence of ethical, social, and legal concerns due to the rapid pace of advancements.
To foster interdisciplinary discussions on these far-reaching implications, Nature Machine Intelligence serves as a platform for dialogue facilitated through Comments, News Features, News & Views articles, and Correspondence. Our goal is to encourage a comprehensive examination of these subjects.
Similar to all Nature-branded journals, Nature Machine Intelligence operates under the guidance of a team of skilled editors. We adhere to a fair and rigorous peer-review process, ensuring high standards of copy-editing and production, swift publication, and editorial independence.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


