DNA hypomethylation promotes UHRF1-and SUV39H1/H2-dependent crosstalk between H3K18ub and H3K9me3 to reinforce heterochromatin states
IF 14.5
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
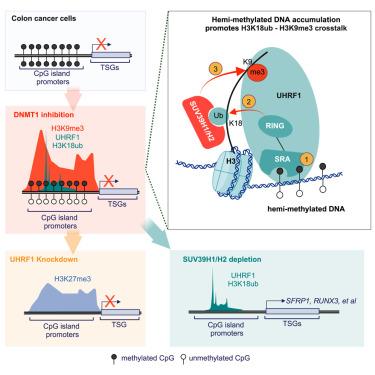
Mono-ubiquitination of lysine 18 on histone H3 (H3K18ub), catalyzed by UHRF1, is a DNMT1 docking site that facilitates replication-coupled DNA methylation maintenance. Its functions beyond this are unknown. Here, we genomically map simultaneous increases in UHRF1-dependent H3K18ub and SUV39H1/H2-dependent H3K9me3 following DNMT1 inhibition. Mechanistically, transient accumulation of hemi-methylated DNA at CpG islands facilitates UHRF1 recruitment and E3 ligase activity toward H3K18. Notably, H3K18ub enhances SUV39H1/H2 methyltransferase activity and, in colon cancer cells, nucleates new H3K9me3 domains at CpG island promoters of DNA methylation-silenced tumor suppressor genes (TSGs). Disrupting UHRF1 enzyme activity prevents H3K9me3 accumulation while promoting PRC2-dependent H3K27me3 as a tertiary layer of gene repression in these regions. By contrast, disrupting H3K18ub-dependent SUV39H1/H2 activity enhances the transcriptional activating and antiproliferative effects of DNMT1 inhibition. Collectively, these findings reveal roles for UHRF1 and H3K18ub in regulating a hierarchy of repressive histone methylation signaling and rationalize a combination strategy for epigenetic cancer therapy.
求助全文
约1分钟内获得全文
求助全文
来源期刊

Molecular Cell
生物-生化与分子生物学
CiteScore
26.00
自引率
3.80%
发文量
389
审稿时长
1 months
期刊介绍:
Molecular Cell is a companion to Cell, the leading journal of biology and the highest-impact journal in the world. Launched in December 1997 and published monthly. Molecular Cell is dedicated to publishing cutting-edge research in molecular biology, focusing on fundamental cellular processes. The journal encompasses a wide range of topics, including DNA replication, recombination, and repair; Chromatin biology and genome organization; Transcription; RNA processing and decay; Non-coding RNA function; Translation; Protein folding, modification, and quality control; Signal transduction pathways; Cell cycle and checkpoints; Cell death; Autophagy; Metabolism.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


