Binding interaction of food preservatives with glycosylated DNA: In vitro assessment and computational studies
IF 4
2区 化学
Q2 CHEMISTRY, PHYSICAL
引用次数: 0
Abstract
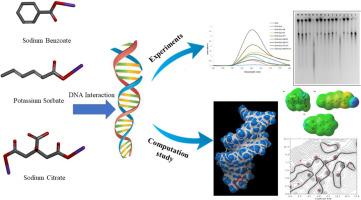
This work aims to evaluate the interaction between sodium benzoate (SB), potassium sorbate (PS), and sodium dihydrogen citrate (CIT) as food preservatives, individually, as well as in combinations of two and three, with DNA and glycosylation DNA to determine the allowable amount of food additive consumption to mitigate potential harm to human health. Various techniques including UV-Vis absorption, fluorescence spectroscopy, DNA amadori product formation, and agarose gel electrophoresis, were employed to observe these interactions. Furthermore, pioneering molecular docking and molecular dynamics simulations were performed to gain deeper insights into the interactions between these preservative compounds and DNA. It is also noteworthy that there have been no documented biological or computational studies concerning citrate. The results revealed that all three food preservatives could interact with DNA, leading to conformational changes as indicated by increased absorption and fluorescence intensity. Both SB and PS were found to damage the DNA structure and promote the formation of DNA Amadori products. Moreover, all treatment groups exhibited a decrease in migration speed on the agarose gel, suggesting binding to DNA and alterations in the supercoiled form of DNA structure. Interestingly, the groups treated with citrate showed a smeared appearance, DNA strand fragmentation, and cleavage. Molecular docking complemented the in vitro achievement that SB, PS, and CIT could interact with DNA. DFT analysis showed that CIT exhibited the best thermodynamic and kinetic stability compared to other food additives.
食品防腐剂与糖基化DNA的结合相互作用:体外评估和计算研究
本研究旨在评估苯甲酸钠(SB)、山梨酸钾(PS)和柠檬酸二氢钠(CIT)作为食品防腐剂与DNA和糖基化DNA的相互作用,以及两者和三者的组合,以确定食品添加剂的允许用量,以减轻对人类健康的潜在危害。利用紫外-可见吸收、荧光光谱、DNA amadori产物形成和琼脂糖凝胶电泳等多种技术来观察这些相互作用。此外,进行了开创性的分子对接和分子动力学模拟,以更深入地了解这些防腐化合物与DNA之间的相互作用。同样值得注意的是,目前还没有关于柠檬酸盐的生物学或计算研究。结果表明,这三种食品防腐剂都可以与DNA相互作用,导致构象变化,这表明吸收和荧光强度增加。发现SB和PS都能破坏DNA结构,促进DNA Amadori产物的形成。此外,所有处理组在琼脂糖凝胶上的迁移速度都有所下降,这表明与DNA的结合以及DNA超螺旋结构的改变。有趣的是,用柠檬酸处理的组表现出涂污的外观,DNA链断裂和切割。分子对接补充了SB、PS和CIT与DNA相互作用的体外成果。DFT分析表明,与其他食品添加剂相比,CIT具有最好的热力学和动力学稳定性。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Journal of Molecular Structure
化学-物理化学
CiteScore
7.10
自引率
15.80%
发文量
2384
审稿时长
45 days
期刊介绍:
The Journal of Molecular Structure is dedicated to the publication of full-length articles and review papers, providing important new structural information on all types of chemical species including:
• Stable and unstable molecules in all types of environments (vapour, molecular beam, liquid, solution, liquid crystal, solid state, matrix-isolated, surface-absorbed etc.)
• Chemical intermediates
• Molecules in excited states
• Biological molecules
• Polymers.
The methods used may include any combination of spectroscopic and non-spectroscopic techniques, for example:
• Infrared spectroscopy (mid, far, near)
• Raman spectroscopy and non-linear Raman methods (CARS, etc.)
• Electronic absorption spectroscopy
• Optical rotatory dispersion and circular dichroism
• Fluorescence and phosphorescence techniques
• Electron spectroscopies (PES, XPS), EXAFS, etc.
• Microwave spectroscopy
• Electron diffraction
• NMR and ESR spectroscopies
• Mössbauer spectroscopy
• X-ray crystallography
• Charge Density Analyses
• Computational Studies (supplementing experimental methods)
We encourage publications combining theoretical and experimental approaches. The structural insights gained by the studies should be correlated with the properties, activity and/ or reactivity of the molecule under investigation and the relevance of this molecule and its implications should be discussed.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


